* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Type III Secretion System
Survey
Document related concepts
Extracellular matrix wikipedia , lookup
Cell membrane wikipedia , lookup
SNARE (protein) wikipedia , lookup
Cell nucleus wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Magnesium transporter wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein moonlighting wikipedia , lookup
Endomembrane system wikipedia , lookup
Signal transduction wikipedia , lookup
Bacterial microcompartment wikipedia , lookup
Protein–protein interaction wikipedia , lookup
List of types of proteins wikipedia , lookup
Western blot wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Transcript
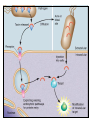
Type III Secretion System Type III Secretion System (TTSS) is a mechanism used by bacteria to establish an infection or symbiotic relationship with a eukaryotic cell by mediating the injection of effector proteins into the hosts cells cytoplasm TTSS injects (translocates) proteins into the cytosol of eukaryotic cells TTSS’s are activated by bacterial contact with host cell surfaces Translocated proteins facilitate bacterial pathogenesis by specifically interfering with host cell signal transduction and other cellular processes E.g. the injected proteins can promote bacterial internalization by mammalian cells in Salmonella and Shigella Macrophage apoptosis in Yersinia spp The type III secretion apparatus is composed of approximately 20 proteins, most of which are located in the inner membrane A subfamily of proteins that include InvG from Salmonella, YscC from Yersinia and MxiD from Shigella are located in the outer membrane and are required for TTSS These proteins are very similar to Secretins that are used to mediate transport across the outer membrane of large molecules • • This ring of helices is a model of the molecular needle of type III secretion system. The model is a combination of the crystal structure of the single subunit and 3D reconstruction of the needle from electron microscopy. •Most of the inner membrane proteins are homologous to components of the flagellar biosynthesis apparatus FliK is a protein is used for flagella construction that signals the completion of the hook component. Once the hook is completed proteins that will make the flagella components will then be secreted. TTSS has a FliK homolog in animal pathogens such as Salmonella and Shigella. The FliK homolog senses when the needle structure is completed and can send a signal(s) to the secretion system so that proteins needed for the needle secretion will stop being produced Proteins which constitute the type III secretion apparatus are conserved among different pathogens. This suggest that the genes have been spread by horizontal gene transfer Therefore, gene clusters are either contained on plasmids or pathogenicity islands •TTSS is coded for by a plasmid called pIB1 in Yersinia spp. Pathogenicity island of E coli O157:H7 genomes The effector genes are not linked between species showing that they are independent of the genes for TTSS protein secretion This allows the bacteria to adapt to host countermeasures or to a new host This is an important process because each different type of bacteria has a preferential niche, which requires different effector proteins Proteins that are predestined for transport through a membrane must be prevented from assuming their three dimensional shape prior to transport Proteins in their assembled active form are too large to pass through the small opening of the needle structure The Role of Chaperones Structurally conserved chaperones which specifically bind to individual secreted proteins are important in TTSS by preventing premature interactions of the secreted factors with other proteins. Chaperones also ensure presecretory stabilization and efficient secretion Lack of specific chaperones can reduce the secretion of the protein due to degradation in the bacteria cytoplasm E.g. In shigella the IpgC chaperone binds to IpaB and IpaC and inhibits premature association and degradation of the IpaBC protein complex before it is secreted Chaperones cap the region required for translocation to prevent premature interaction with other proteins in TTSS apparatus or from self-aggregation prior to secretion The region of the protein that is bound to the chaperone is unfolded and resistant to proteolysis The unbound C terminus remains active Not all proteins require a chaperone However, proteins with chaperones are secreted more rapidly If the protein has no chaperone it is generally not as efficient as those that do have a chaperone associated with it Summary Points TTSS uses a needle like structure to move proteins from the bacteria to the eukaryotic host across all three membranes Many of the proteins that form TTSS are homologous to those found in bacteria flagella Show support that flagella and TTSS are formed in a similar fashion Genes that code for TTSS are highly conserved between different species; however the genes for effector proteins are different in each species Shows that the proteins that form TTSS are independent from effector proteins Chaperones prevent degradation of effector proteins, early interaction with other proteins, and ensure a higher rate of translocation