* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Transcription and the Central Dogma
Gel electrophoresis of nucleic acids wikipedia , lookup
Genetic code wikipedia , lookup
Transcription factor wikipedia , lookup
Gene regulatory network wikipedia , lookup
Molecular cloning wikipedia , lookup
Community fingerprinting wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Molecular evolution wikipedia , lookup
RNA interference wikipedia , lookup
Real-time polymerase chain reaction wikipedia , lookup
Biosynthesis wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Non-coding DNA wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Polyadenylation wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Messenger RNA wikipedia , lookup
RNA silencing wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Gene expression wikipedia , lookup
Transcriptional regulation wikipedia , lookup
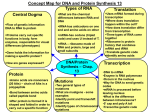
Transcription and the Central Dogma 1 • 1961: DNA is the molecule which stores genetic information, but – DNA is in nucleus, ribosomes (where protein synthesis takes place) are in the cytoplasm. – RNA is synthesized in the nucleus, is similar to DNA. – RNA migrates to cytoplasm (where ribosomes are) – Amount of RNA generally proportional to amount of proteins in the cell. • All this suggests role of RNA as messenger. 2 About RNA 1) DNA is double stranded, but RNA is single stranded. However, RNA can base-pair with itself to create double stranded regions. RNA DNA tRNA genetics.gsk.com/graphics/ dna-big.gif http://www.fhi-berlin.mpg.de/th/JG/RNA.jpg http://www.santafe.edu/images/rna.gif About RNA-2 2) RNA contains ribose instead of deoxyribose 3) RNA contains uracil instead of thymine. www.layevangelism.com/.../ deoxyribose.htm http://www.rothamsted.bbsrc.ac.uk/notebook/courses/guide/images/uracil.gif 3 4 3 kinds of RNA mRNA: a copy of the gene; is translated to make protein. tRNA: smallest RNA, does actual decoding. tRNA rRNA: 3 sizes that, along with proteins, make up a ribosome. rRNA http://www.cu.lu/labext/rcms/cppe/traducti/tjpeg/trna.jpeg; Tobin and Duschek, Asking About Life; http://www.tokyo-ed.ac.jp/genet/mutation/nort.gif More kinds of RNA 5 • snRNAs – Small RNAs (100-200 bases) that attach to proteins to form snRNPs (small nuclear ribonucleoproteins) – snRNPs are components of the spliceosome • Removes introns from pre-mRNA • snoRNAs – Small nucleolar RNAs, process rRNA – Modify other RNAs; guide RNA of telomerase • Other RNAs, e.g. Xist • All RNA comes from the DNA (RNA “genes”) Transcription: making mRNA • RNA a polymer assembled from monomers – Ribonucleoside triphosphates: ATP, UTP, GTP,CTP • RNA polymerase – Multi-component enzyme – Needs a template, but NOT a primer – A component (sigma in bacteria) recognizes the promoter as the place on DNA to start synthesis – Synthesis proceeds 5’ to 3’, just as in DNA • mRNA is complementary and antiparallel to the DNA strand being copied. 6 Transcription-2 7 • The order of nucleotides in the RNA reflects the order in the DNA • If RNA is complementary to one DNA strand, then it is identical (except for T change to U) to the other DNA strand. Either DNA strand may contain the gene! Transcription just runs the other direction. Sense, antisense Compare the sense strand of the DNA to the mRNA. Note that mRNA synthesis will be 5’ to 3’ and antiparallel. http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/S/SenseStrand.gif 8 More about RNA polymerase 9 • In bacteria, components are ββ’α2φσ • RNA polymerase is processive; once enzyme attaches to DNA, it can copy >10,000 nucleotides without falling off. • In eukaryotes, there are 3 RNA polymerases: – One for rRNA – One for tRNAs and some rRNA – One for all mRNAs and some small RNAs (involved in RNA processing) Transcription needs a Promoter 10 A promoter is nontranscribed DNA Prokaryotes Eukaryotes http://opbs.okstate.edu/~petracek/2002%20Gene%20expression/img043.gif The Process of Transcription 11 • Promoter recognition: 2 consensus sequences – The -10 region: TATAAT (10 bases upstream from where transcription actually starts. – The -35 region, farther upstream, also important. – “Consensus” sequence meaning the DNA sequence from many genes averages out to this. – The closer these 2 regions actually are to the consensus sequences, the “stronger” the promoter, meaning the more likely RNA polymerase binding and transcription will occur. Consensus sequence Numbers indicate the percentage of different genes in which that nucleotide appears in that spot in the promoter sequence. http://www.uark.edu/campus-resources/mivey/m4233/promoter.gif 12 The Process of Transcription-2 13 • After binding to the promoter, polymerase “melts” DNA, lines up first base at the +1 site = Initiation. • RNA synthesis continues (Elongation), only the template strand being transcribed. • Termination: must be a stop sign, right? – In bacteria, hairpin loop followed by run of U’s in the RNA. Of course, the DNA must code for complementary bases and a run of A’s. See next. – Termination factor “rho”. Accessory protein. Termination of Transcription in Bacteria In euks, termination occurs with a processing step. 14 The hairpin loop destabilizes the interactions between the DNA, mRNA, and polymerase; U-A basepairs are very weak, and the complex falls apart. http://www.blc.arizona.edu/marty/411/Modules/Weaver/Chap6/Fig.0649ac.gif About mRNA structure, etc. 15 • Start site of transcription is NOT equal to start site of Translation – First codon read, AUG, is downstream from the first ribonucleotides. +1 is transcription start, not translation start. – AUG marks the beginning of an Open Reading Frame (ORF). • Lifetime of a eukaryotic mRNA is variable • For prokaryotes, mRNA is short lived, fits in with need of microbes to respond quickly to changes in environment. Eukaryotic transcription • Occurs in nucleus, then mRNA goes to cytoplasm • Promoters but also enhancers – Enhancers also segments of DNA • Eukaryotic RNA requires processing – Pre-mRNAs found in nucleus as hnRNA • Heterogeneous nuclear RNA • With proteins: hnRNPs: heterogeneous nuclear ribonucleoprotein particles 16 Initiation of transcription (euks) • Goldberg-Hogness (TATA) box at -35 • CAAT box at -80 (non specific, but important) – GGCCAATCT consensus sequence • Enhancers – Elements of DNA that promote transcription – Can be upstream, downstream, even in gene 17 Eukaryotic RNA polymerase 18 • A 10 subunit machine • Makes several attempts, abortive transcription • Key: successful synthesis of short DNA-RNA hybrid, stabilizes association of Pol with DNA – Then, processive to a high degree, transcribes until knocked off by termination signal. Processing of mRNA • Cap – 7-methyl guanosine is added as a cap, 5’ to 5’ – Cap aids in binding of mRNA to the ribosome – Shine-Delgarno seq. does same in prokaryotes http://www.blc.arizona.edu/Marty/429/Lectures/Figures/CAP.GIF 19 Processing of mRNA-2 • Poly-A tail – In several steps, end of mRNA is cleaved off, and several rounds of AAAAAAA are added – Poly-A tail improves stability of mRNA, resists degradation by nucleases in cell. departments.oxy.edu/.../ processing_of_hnrnas.htm 20 21 Introns and Exons •Introns are intervening sequences that do not contain information for making the protein. Exons are the coding sequences left behind. http://www.emc.maricopa.edu/faculty/farabee/BIOBK/exintrons.gif Removal of Introns • Three types of intron-removal mechanisms – In nucleus, hnRNA (heterogeneous nuclear RNA) • Made of precursor mRNA and snRNPs – snRNPs: small nuclear ribonucleoproteins • Aggregated into particles called spliceosomes – In some systems, intron codes for protein that splices out the intron! – In Tetrahymena, intron is self-splicing: ribozyme 22 Splicing of Introns • In most systems, snRNPs cut mRNA, introns come out, and snRNPs help splice exons back together. • Particular sequences in the mRNA mark the beginning, end of exons and introns so snRNPs can do their job. http://www.plantsci.cam.ac.uk/Haseloff/SITEGRAPHICS/splice1.GIF 23 Introns: 24 How were they discovered? RNA-DNA hybrids weren’t colinear- loops of DNA extend out where there is no RNA to base pair with it. RNA = red. Mutations in introns: don’t have much affect unless: •Mutation is near a splice site •Mutation is in a regulatory region (which could be in an intron) •There is a separate gene within the intron. Why are there introns? 25 • Very ancient? – Located in the same positions in genes common to plants and animals. Maybe bacteria had them once and lost them. – Self-splicing RNAs may be related to RNA as the first nucleic acid, a popular idea in evolution. • Exon-shuffling: a model for gene evolution – Some proteins fold into connected, functional sections called domains; these correspond to exons – Perhaps exons were copied, shuffled to create new genes. Several human genes share exons. Relationship between protein domains and exons 26 http://www.mrclmb.cam.ac.uk/genomes/cvogel/SupraDomains/Data_new/sd.figure.supra-domain.jpg