* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Phenotype to genotype (Top down)
Pharmacogenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Medical genetics wikipedia , lookup
Genetic engineering wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Designer baby wikipedia , lookup
Genetic code wikipedia , lookup
Behavioural genetics wikipedia , lookup
Adaptive evolution in the human genome wikipedia , lookup
Genetic testing wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Public health genomics wikipedia , lookup
Koinophilia wikipedia , lookup
Dual inheritance theory wikipedia , lookup
Genome (book) wikipedia , lookup
Heritability of IQ wikipedia , lookup
History of genetic engineering wikipedia , lookup
Human genetic variation wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Genetic drift wikipedia , lookup
Group selection wikipedia , lookup
Quantitative trait locus wikipedia , lookup
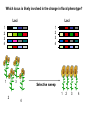
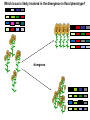
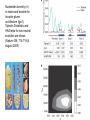
Plant of the day! Pebble plants, Lithops, dwarf xerophytes Aizoaceae South African Plants consist of one or more pairs of bulbous leaves – almost no stem Leaf markings appear to help plant match its background and be less vulnerable to herbivory Lithops lesliei Adaptation Goals • Understand some top down and bottom up approaches used to identify genes responsible for adaptations • Explain patterns of sequence variation expected with positive and balancing selection • Understand the principles of population genetic tests of selection The genetic basis of adaptation • Phenotype to genotype (Top down) – Candidate loci for traits responsible for phenotypic differences – QTL, association mapping • Genotype to phenotype (Bottom up) – Population genetics Which locus is likely involved in the change in floral phenotype? Loci Loci 1 2 3 4 1 2 3 4 1 3 2 Selective sweep 1 4 2 3 4 Which locus is likely involved in the divergence in floral phenotype? divergence Quantitative trait loci (QTL) are regions of the genome associated with the trait in question -May not be the loci in other individuals/environments -Statistical issues (sample size, genes of small effect, epistasis) -Can be large regions of a chromosome (further mapping in region needed) -Can't perform in all species Rodney Mauricio 2001 Association mapping Associations between markers (SNPs) and phenotypes in natural populations allele1 allele2 SNP1 100% 0% SNP2 50% 50% allele1 allele2 SNP1 0% 100% SNP2 50% 50% Can find precise mutation responsible No crossing required False associations due to population structure Large sample size, many markers needed (if no candidate loci) The genetic basis of adaptation • Phenotype to genotype (Top down) – Candidate loci for traits responsible for phenotypic differences – QTL, association mapping • Genotype to phenotype (Bottom up) – Population genetics Neutral Theory Kimura (1968, 1983) Motoo Kimura (1924-1994) Ph.D. University of Wisconsin in 1956 Under James Crow Kimura argued that the great majority of evolutionary changes at the molecular level are not caused by selection but by random genetic drift. Neutral Theory: Evidence Molecular evolution takes place at a relatively constant rate, simply through random genetic drift, enough to provide a “molecular clock” of evolution. Selection-Neutral Debate • Kimura’s work spawned a heated debate on the relative importance of neutral evolution (genetic drift) versus genetic variation that is a result of natural selection. Neutral Theory • Now considered the “null model” against which evidence for selection should be tested Detecting Natural Selection There are many statistical tests for detecting Natural selection The approach is to test for deviations from a null neutral model (where genetic variation arises only from genetic drift) Null hypothesis: Neutral, no selection Deviation from Neutral: selection Inferences regarding selection provide a powerful tool for the prediction of genes underlying traits of ecological or economic value The effects of selection on the genome Directional selection – Best allele(s) sweep to fixation – Loss of variation – Change in frequency distribution of polymorphisms – Increase in linkage disequilibrium around the site The effects of selection on the genome Balancing selection – Maintains variation that otherwise would be lost to drift – Heterozygote advantage, frequency dependent selection, fluctuating selection, (divergent selection) •A beneficial allele arises •Variants with this allele rapidly spread through the species •Genetic diversity is reduced around this adaptive locus ancestral After selection Chance of detecting positive selection depends on: Time Strength of selection Recombination, mutation Initial frequency Methods for Detecting Selection: A. MacDonald-Kreitman Type Tests B. Site Frequency Spectrum Approaches C. Linkage Disequilibrium (LD) and Haplotype Structure D. Population Differentiation: Lewontin-Krakauer Methods These tests could be applied to single genes, or across the whole genome. Codon Bias in Amino Acid Substitutions • Synonymous substitutions: Mutations that do not cause amino acid change (usually 3rd position) “silent substitutions” • Nonsynonymous substitutions: Mutations that cause amino acid change (1st, 2nd position) “replacement substitutions” QuickTime™ and a Photo - JPEG decompressor are needed to see this picture. A. MacDonald-Kreitman Type Tests (1) Ka/Ks Test Nonsynonymous substitutions Synonymous substitutions Ka Ks >1 • Need coding sequence (sequence that codes proteins) • Ks is used here as the “control”, proxy for neutral evolution so Ka/Ks = 1 neutral evolution • A larger nonsynonymous substitution rate (Ka) than synonymous (Ks) is used as an indication of selection (Ka/Ks >1) • Ka/Ks < 1 ? dN vs. dS for pairwise sequence comparisons in Physalis (nightshade) Richman A D , Kohn J R PNAS 1999;96:168-172 B. Site Frequency Spectrum • Selection affects the distribution of alleles within populations • Method examines site frequency spectrum and compares to neutral expectations • Could be applied to a single locus. Now used often for genomic scans for selective sweeps •Domestication alleles (corn, rice) Nucleotide diversity (π) in maize and teosinte for teosinte glume architecture (tga1). Tajima's D-statistic and HKA tests for non-neutral evolution are shown (Nature 436, 714-719 (4 August 2005) C. Linkage Disequilibrium (LD) • The nonrandom association of alleles from different loci, where they are found more or less frequently together than expected • Selection increases levels of linkage disequilibrium during the process of selection D. Population Differentiation: Lewontin-Krakauer Methods • Selection would often increase the degree of genetic distance between populations • Compute pairwise genetic distances (FST) for many loci between populations • When a locus shows extraordinary levels of genetic distance relative to other loci, this locus is a candidate for positive selection Example of Fst scan in sunflower cM