* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Compressed Adjacency Matrices: Untangling Gene Regulatory
Survey
Document related concepts
Transcript
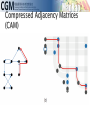
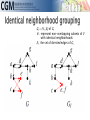
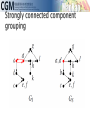
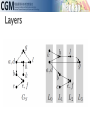
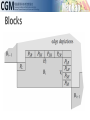
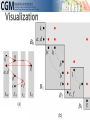
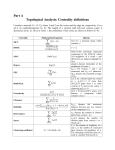
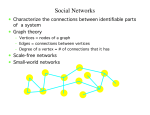
infoVis 2012 Kasper Dinkla, Michel A. Westenberg, and Jarke J. van Wijk Introduction Related work ◦ Node-Link Based Representations ◦ ZAME: Interactive Large-Scale Graph Visualization TimeMatrix User Study Conclusion Gene Regulatory Network (GRN) ◦ Low in-degree Every gene is regulated by only a few other genes. Therefore, all nodes of the network have a low indegree. ◦ Scale-free out-degree There are few genes that regulate many others, and many genes that regulate few others. Therefore, the network’s out-degree distribution follows a power law. ◦ Few cycles The network has few cycles because genes rarely (indirectly) regulate each other both ways. node-link diagrams (low edge to node ratio) ◦ high number of intersecting edges adjacency matrices (high edge to node ratio) ◦ space-inefficient, sparse network green: promotion red: inhibition orange: both blue: unspecified Compressed Adjacency Matrices (CAM) ◦ Compactness This enables a detailed overview of the entire network. ◦ Localization of motifs This enables quick detection of subnetworks of interest. ◦ Consistent arrangement This facilitates interaction while preserving visual orientation. the conversion of a network to a CAM is not trivial and consists of six steps: 1. the network is decomposed into weakly connected components 2. nodes with identical neighborhoods are grouped 3. strongly connected components are detected and grouped to form a DAG 4. the nodes of the DAG are partitioned into layers 5. the layers are turned into blocks that form the backbone of the CAM 6. the blocks are concatenated to form a cascade from which node positions directed graph G = (V,E) V : the set of vertices (nodes) E : the set of directed edges between vertices of G GI = (VI ,EI) of G, VI : represent non-overlapping subsets of V with identical neighborhoods EI: the set of directed edges of GI Layers map directly to blocks ◦ a block Bi is derived from its corresponding layer Li Hi and Vi, that specify the horizontal and vertical ordering of Li’s vertices in the CAM partitioning the vertices of Li into five classes: Leaf (PL) Short root (PSR) Long root (PLR) Short hub (PSH) Long hub (PLH) ◦ Vertex in Li without successors ◦ Vertex in Li that has a successor but no predecessors and all successors are leaves in Li+1 ◦ Vertex in Li that has a successor but no predecessors and is not a short root ◦ Vertex in Li that has a predecessor and successor, and all successors are leaves in Li+1 ◦ Vertex in Li that has a predecessor and successor, but is not a short hub categorizing visual analytic tasks in temporal social network analysis (Tasks 1, 2, and 3), proposing an adjacency-matrix-based visual representation (TimeMatrix) for analyzing temporal graphs that complement node-link temporal graph visualization techniques, and supplementing TimeMatrix with interaction techniques supporting highly interactive visual exploration of real-world social networks across multiple levels of analysis.