Automate Function Prediction
... (http://genome.cshlp.org/content/early/2011/07/22/gr.104687.109) ...
... (http://genome.cshlp.org/content/early/2011/07/22/gr.104687.109) ...
PROTEOME:
... • Label protein samples with heavy and light reagent • Reagent contains affinity tag and heavy or light isotopes Chemically reactive group: forms a covalent bond to the protein or peptide Isotope-labeled linker: heavy or light, depending on which isotope is used Affinity tag: enables the protein or ...
... • Label protein samples with heavy and light reagent • Reagent contains affinity tag and heavy or light isotopes Chemically reactive group: forms a covalent bond to the protein or peptide Isotope-labeled linker: heavy or light, depending on which isotope is used Affinity tag: enables the protein or ...
Protein Synthesis: Comprehesive Review PowerPoint Slides
... – One ribosome can assemble protein of 400 amino acids in 20 seconds – 300,000 identical mRNA molecules may be undergoing simultaneous ...
... – One ribosome can assemble protein of 400 amino acids in 20 seconds – 300,000 identical mRNA molecules may be undergoing simultaneous ...
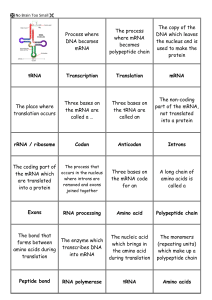
Gene expression flash cards
... the mRNA which occurs in the nucleus where introns are are translated removed and exons into a protein joined together ...
... the mRNA which occurs in the nucleus where introns are are translated removed and exons into a protein joined together ...
Gene Section chromosome 18-like 1) Atlas of Genetics and Cytogenetics
... In the SS18L1/ SSX1 transcript detected in the synovial sarcoma, the exon 10 of SS18L1, which corresponds to exon 10 of SS18, was fused to exon 6 of SSX1. Abnormal protein In the putative SS18L1/SSX1 chimeric protein, the last 8 amino acid residues of the SS18L1 protein are replaced by 78 amino acid ...
... In the SS18L1/ SSX1 transcript detected in the synovial sarcoma, the exon 10 of SS18L1, which corresponds to exon 10 of SS18, was fused to exon 6 of SSX1. Abnormal protein In the putative SS18L1/SSX1 chimeric protein, the last 8 amino acid residues of the SS18L1 protein are replaced by 78 amino acid ...
When it comes to reliable automation of protein digestion for LC and
... protein digestion followed by sample preparation for LC-MS, the choice is simple. The ProPrep LC is the only instrument that has been specifically designed for this purpose. The ProPrep LC gives you peace of mind and your samples the best treatment. The ProPrep LC comes standard with one reaction bl ...
... protein digestion followed by sample preparation for LC-MS, the choice is simple. The ProPrep LC is the only instrument that has been specifically designed for this purpose. The ProPrep LC gives you peace of mind and your samples the best treatment. The ProPrep LC comes standard with one reaction bl ...
Organic Macromolecules Cloze Worksheet
... Proteins are macromolecules that consist of long, unbranched chains of amino acids. These chains may contain about 20 up to hundreds of acids. An example of the size of proteins is the red pigment in red blood cells called haemoglobin with the chemical formula – C3032 H4816 O872 N780 S8 Fe4 Each cel ...
... Proteins are macromolecules that consist of long, unbranched chains of amino acids. These chains may contain about 20 up to hundreds of acids. An example of the size of proteins is the red pigment in red blood cells called haemoglobin with the chemical formula – C3032 H4816 O872 N780 S8 Fe4 Each cel ...
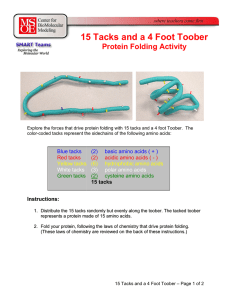
15 Tacks and a 4 Foot Toober
... After everyone has folded their toober as best they can, the teacher can point out: • Every toober had a different random sequence of tacks (amino acids) and therefore each toober (protein) folded into a different structure. • Some sequences of tacks were more easily folded into a reasonable structu ...
... After everyone has folded their toober as best they can, the teacher can point out: • Every toober had a different random sequence of tacks (amino acids) and therefore each toober (protein) folded into a different structure. • Some sequences of tacks were more easily folded into a reasonable structu ...
CFTR – gene cloning and initial bioinformatic analysis Riordan et 12
... Whole protein is two similar halves each with 6 membrane Spanning domains (hydropathic peaks) and two NBFs (hydrophilic regions) and a charged R region ...
... Whole protein is two similar halves each with 6 membrane Spanning domains (hydropathic peaks) and two NBFs (hydrophilic regions) and a charged R region ...
Supporting text S1
... the tryptophan binding pocket. In B. stearothermophilus TRAP, Leu24 interacts with Leu44 from the neighboring chain stabilizing the hydrophobic cluster, while residues Leu24 and Ile44 play the same role in B. subtilis TRAP. In B. halodurans TRAP the corresponding residues are Met24 and Met44, respec ...
... the tryptophan binding pocket. In B. stearothermophilus TRAP, Leu24 interacts with Leu44 from the neighboring chain stabilizing the hydrophobic cluster, while residues Leu24 and Ile44 play the same role in B. subtilis TRAP. In B. halodurans TRAP the corresponding residues are Met24 and Met44, respec ...
ppt - UCLA Chemistry and Biochemistry
... • Preferential binding of transition state: binding interactions between the enzyme and TS are maximized; they are greater than those in the enzyme-substrate or enzyme-product complexes • General acid and general base catalysis: functional groups of the enzyme donate &/or accept protons • Covalent c ...
... • Preferential binding of transition state: binding interactions between the enzyme and TS are maximized; they are greater than those in the enzyme-substrate or enzyme-product complexes • General acid and general base catalysis: functional groups of the enzyme donate &/or accept protons • Covalent c ...
Ch4Carbonand5Macromolecules
... • Amino acids are linked together by condensation to form polypeptides. • There are 20 different amino acids in polypeptides synthesized on ribosomes. • Amino acids can be linked together in any sequence giving a huge range of possible polypeptides. • The amino acid sequence of polypeptides is coded ...
... • Amino acids are linked together by condensation to form polypeptides. • There are 20 different amino acids in polypeptides synthesized on ribosomes. • Amino acids can be linked together in any sequence giving a huge range of possible polypeptides. • The amino acid sequence of polypeptides is coded ...
xcjkhfk
... Fish oil beneficial anti-inflammatory Early Studies : Good Safety profile Clin Nutr 2013;32:224 JPEN 2012; 36:81S ...
... Fish oil beneficial anti-inflammatory Early Studies : Good Safety profile Clin Nutr 2013;32:224 JPEN 2012; 36:81S ...
Lecture 6 mRNA splicing and protein synthesis
... and may contain non-Watson-Crick base pairs, e.g. GU. This portion of the tRNA is called the acceptor since the amino acid is carried by the tRNA while attached to the 3'-terminal OH group. 4. have a D loop and a TpsiC loop. 5. have an anti-codon loop. 6. terminate at the 3'-end with the sequence 5' ...
... and may contain non-Watson-Crick base pairs, e.g. GU. This portion of the tRNA is called the acceptor since the amino acid is carried by the tRNA while attached to the 3'-terminal OH group. 4. have a D loop and a TpsiC loop. 5. have an anti-codon loop. 6. terminate at the 3'-end with the sequence 5' ...
Chapter 2
... • Primary structure – Sequence of amino acids – In large part determines other protein features ...
... • Primary structure – Sequence of amino acids – In large part determines other protein features ...
Comparative Proteomics Kit I: Protein Profiler Module
... • Can be completed in three 45 minute lab sessions ...
... • Can be completed in three 45 minute lab sessions ...
Translation - Fog.ccsf.edu
... One of the three STOP codons mark the end of translation The stop codons are recognized by proteins known as release factors that do not specify any amino acids The release factor triggers an addition of water to the end of the polypeptide chain the release of the new protein. ...
... One of the three STOP codons mark the end of translation The stop codons are recognized by proteins known as release factors that do not specify any amino acids The release factor triggers an addition of water to the end of the polypeptide chain the release of the new protein. ...
Bio301 Biochemistry I
... Where ci is the molar concentration of the ith ionic species and Zi is its ionic charge. At high ionic strengths the solubilities ofproteins as well as those of most other substances, decrease. This effect is known as salting out. You have given 1.0 M solutions of NaCl, (NH4) 2SO4 and K3PO4.In which ...
... Where ci is the molar concentration of the ith ionic species and Zi is its ionic charge. At high ionic strengths the solubilities ofproteins as well as those of most other substances, decrease. This effect is known as salting out. You have given 1.0 M solutions of NaCl, (NH4) 2SO4 and K3PO4.In which ...
general western blot troubleshooting tips
... Filter the secondary with a 0.2 µm filter to remove any aggregates. ...
... Filter the secondary with a 0.2 µm filter to remove any aggregates. ...
Mistakes Happen
... A point mutation: any mutation in which one base of the gene sequence is changed. A single base can be inserted, deleted or substituted. (you may see it used synonymously with just substitution, but the official definition is broader) Example: Typing the word “Mog” or “Doog” when you wanted to type ...
... A point mutation: any mutation in which one base of the gene sequence is changed. A single base can be inserted, deleted or substituted. (you may see it used synonymously with just substitution, but the official definition is broader) Example: Typing the word “Mog” or “Doog” when you wanted to type ...
File - Mr. Shanks` Class
... Keratin (in hair) is an alpha-helix Fibroin (in spider web silk) is a beta-pleated sheet ...
... Keratin (in hair) is an alpha-helix Fibroin (in spider web silk) is a beta-pleated sheet ...
Protein structure prediction

Protein structure prediction is the prediction of the three-dimensional structure of a protein from its amino acid sequence — that is, the prediction of its folding and its secondary, tertiary, and quaternary structure from its primary structure. Structure prediction is fundamentally different from the inverse problem of protein design. Protein structure prediction is one of the most important goals pursued by bioinformatics and theoretical chemistry; it is highly important in medicine (for example, in drug design) and biotechnology (for example, in the design of novel enzymes). Every two years, the performance of current methods is assessed in the CASP experiment (Critical Assessment of Techniques for Protein Structure Prediction). A continuous evaluation of protein structure prediction web servers is performed by the community project CAMEO3D.