The Genome-linked Protein of Picornaviruses. VIII. Complete Amino
... In the results presented here we have conclusively shown the complete primary structure of poliovirus VPg. It is 22 amino acids in length, 2354 Mr, and terminates in -valyl-glutamineCOOH. The nature of the carboxy-terminus did not surprise us since in the poliovirus polyprotein, from which VPg is ul ...
... In the results presented here we have conclusively shown the complete primary structure of poliovirus VPg. It is 22 amino acids in length, 2354 Mr, and terminates in -valyl-glutamineCOOH. The nature of the carboxy-terminus did not surprise us since in the poliovirus polyprotein, from which VPg is ul ...
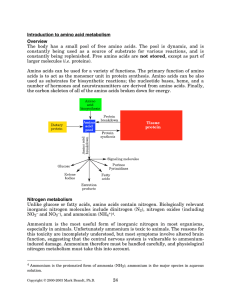
Introduction to amino acid metabolism Overview - Rose
... Two other amino acids, cysteine and tyrosine are typically considered to be nonessential amino acids. This is not really accurate, because each of these can only be synthesized from an essential amino acid. Cysteine is synthesized from methionine, and tyrosine is synthesized from phenylalanine. If t ...
... Two other amino acids, cysteine and tyrosine are typically considered to be nonessential amino acids. This is not really accurate, because each of these can only be synthesized from an essential amino acid. Cysteine is synthesized from methionine, and tyrosine is synthesized from phenylalanine. If t ...
Document
... the molecules in a material. Increasing the temperature increases the translational motion of molecules. ...
... the molecules in a material. Increasing the temperature increases the translational motion of molecules. ...
“Nice” plotting of proteins
... Polynomial fits are sound if higher order derivatives of the “true” function (beyond the order of the polynomial) do not contribute significantly to the description of the function. However, the reverse is also true… If high order derivatives do make a significant contribution then the fit is not so ...
... Polynomial fits are sound if higher order derivatives of the “true” function (beyond the order of the polynomial) do not contribute significantly to the description of the function. However, the reverse is also true… If high order derivatives do make a significant contribution then the fit is not so ...
Molecular and Cellular Biology, December 2001, p
... incubated for 30 min on ice and centrifuged at 16,000 × g for 30 min at 4°C. The supernatant was recovered and centrifuged for 1 h at 100,000 × g, at 4°C (cytoplasmic extract). AntimHDAC6 antibody (37) or peptide-blocked anti-mHDAC6 antibody (antibody preincubated for 30 min at 4°C with its target p ...
... incubated for 30 min on ice and centrifuged at 16,000 × g for 30 min at 4°C. The supernatant was recovered and centrifuged for 1 h at 100,000 × g, at 4°C (cytoplasmic extract). AntimHDAC6 antibody (37) or peptide-blocked anti-mHDAC6 antibody (antibody preincubated for 30 min at 4°C with its target p ...
INDIGO-BINDING DOMAINS IN CELLULASE MOLECULES
... residues seem to play a major role in binding indigo. Indigo is a hydrophobic compound (insoluble in water), and its aromatic rings may interact with the aromatic rings of Tyr, Trp and Phe, as well as with side chains of other non-polar amino acids, via the hydrophobic interactions. Another mechanis ...
... residues seem to play a major role in binding indigo. Indigo is a hydrophobic compound (insoluble in water), and its aromatic rings may interact with the aromatic rings of Tyr, Trp and Phe, as well as with side chains of other non-polar amino acids, via the hydrophobic interactions. Another mechanis ...
7 Fig. 1. "Double-sieve" (two- step subtrate selection - SPring-8
... Therefore, IleRS selects the cognate L-isoleucine only on the basis of the size of its hydrophobic side chain, whereas ValRS selects L-valine by size exclusion at the first step followed by hydrophobicity exclusion at the second step. We previously reported the crystal structure of Thermus thermophi ...
... Therefore, IleRS selects the cognate L-isoleucine only on the basis of the size of its hydrophobic side chain, whereas ValRS selects L-valine by size exclusion at the first step followed by hydrophobicity exclusion at the second step. We previously reported the crystal structure of Thermus thermophi ...
SOD binds cell-adhesive peroxidase - Journal of Cell Science
... surface; it could be released from the membrane with high salt. It was thus concluded that the peroxinectin-binding protein is an extracellular SOD (EC-SOD) and a peripheral membrane protein, presumably kept at the cell surface via ionic interaction with its C-terminal region. This interaction with ...
... surface; it could be released from the membrane with high salt. It was thus concluded that the peroxinectin-binding protein is an extracellular SOD (EC-SOD) and a peripheral membrane protein, presumably kept at the cell surface via ionic interaction with its C-terminal region. This interaction with ...
Docking QM/MM
... we have employed the smallest acceptable QM region consisting of the bound ligand and the backbone of the 3 amino acids of the hinge, with the point charges of the outer MM region being electrically embedded. Where bonds cross the QM and MM interface, the valences were satisfied by hydrogen link ato ...
... we have employed the smallest acceptable QM region consisting of the bound ligand and the backbone of the 3 amino acids of the hinge, with the point charges of the outer MM region being electrically embedded. Where bonds cross the QM and MM interface, the valences were satisfied by hydrogen link ato ...
Similarities between putative transport proteins of plant viruses
... CGMMV and TRV sequences were added, in that order, to the TMVlike group. The FMV sequence was added to the two other caulimoviral sequences and the BMV-CMV pair was aligned with the TSV-AIMV pair. Three groups of aligned sequences resulted: the TMV-like group, the caulimoviral group, and the A1MV-li ...
... CGMMV and TRV sequences were added, in that order, to the TMVlike group. The FMV sequence was added to the two other caulimoviral sequences and the BMV-CMV pair was aligned with the TSV-AIMV pair. Three groups of aligned sequences resulted: the TMV-like group, the caulimoviral group, and the A1MV-li ...
Fluorescence Spectroscopy
... keep in mind that protein unfolding reaction is likely to differ from the mechanism of protein folding. Thus the extrapolation of protein folding based on protein unfolding is quite limited. ...
... keep in mind that protein unfolding reaction is likely to differ from the mechanism of protein folding. Thus the extrapolation of protein folding based on protein unfolding is quite limited. ...
Minimal domain of bacterial phytochrome required for chromophore binding and fluorescence
... a new Cys252 residue became responsible for this function. Our hypothesis was based on the similarity between the position of Cys252 and chromphore-binding Cys residues in the GAF domains of plant and cyanobacterial phytochromes, which utilize reduced tetrapyrrole bilins such as phytochromobilin and ...
... a new Cys252 residue became responsible for this function. Our hypothesis was based on the similarity between the position of Cys252 and chromphore-binding Cys residues in the GAF domains of plant and cyanobacterial phytochromes, which utilize reduced tetrapyrrole bilins such as phytochromobilin and ...
npgrj_nchembio_91 405..407 - The Scripps Research Institute
... compared to that of wild-type (WT) ALDH-1 following transient transfection in COS-7 cells. Consistent with previous studies9, the SE probe was found to label both WT and C303A mutant ALDH-1 but not the E269A mutant (Fig. 2a). Notably, the cysteine-reactive CA probe showed the opposite profile, label ...
... compared to that of wild-type (WT) ALDH-1 following transient transfection in COS-7 cells. Consistent with previous studies9, the SE probe was found to label both WT and C303A mutant ALDH-1 but not the E269A mutant (Fig. 2a). Notably, the cysteine-reactive CA probe showed the opposite profile, label ...
in Graminaceous Plants
... MAs are found only in graminaceous plants, although NA has been detected in every plant so far investigated. Therefore, this amino transfer reaction is the first step in the unique biosynthesis of MAs that has evolved in graminaceous plants. NAAT activity is dramatically induced by Fe deficiency and ...
... MAs are found only in graminaceous plants, although NA has been detected in every plant so far investigated. Therefore, this amino transfer reaction is the first step in the unique biosynthesis of MAs that has evolved in graminaceous plants. NAAT activity is dramatically induced by Fe deficiency and ...
In Silico Identification, Classification And Expression
... tolerance of A.thaliana (Liu et al., 2013; Xu et al., 2012). Among the LHCrelated proteins, the early light-induced proteins were the most studied. These proteins play a key role in photoprotection and abiotic stress response in a large number of species such as A. thaliana (Hutin et al., 2003), Rho ...
... tolerance of A.thaliana (Liu et al., 2013; Xu et al., 2012). Among the LHCrelated proteins, the early light-induced proteins were the most studied. These proteins play a key role in photoprotection and abiotic stress response in a large number of species such as A. thaliana (Hutin et al., 2003), Rho ...
Structure-activity Relationships in Flexible Protein Domains
... The guanine dissociation inhibitors RhoGDI and D4GDI inhibit guanosine 50 -diphosphate dissociation from Rho GTPases, keeping these small GTPases in an inactive state. The GDIs are made up of two domains: a ¯exible N-terminal domain of about 70 amino acid residues and a folded 134-residue C-terminal ...
... The guanine dissociation inhibitors RhoGDI and D4GDI inhibit guanosine 50 -diphosphate dissociation from Rho GTPases, keeping these small GTPases in an inactive state. The GDIs are made up of two domains: a ¯exible N-terminal domain of about 70 amino acid residues and a folded 134-residue C-terminal ...
Validating the Location of Fluorescent Protein
... can be used to enhance the resolution of subcellular protein localization, but similar problems with fixation artifacts are often present with this method (Ripper et al., 2008). A second example is provided by the study of Chow et al. (2008) referred to above. In this study of the Rab-A2 family memb ...
... can be used to enhance the resolution of subcellular protein localization, but similar problems with fixation artifacts are often present with this method (Ripper et al., 2008). A second example is provided by the study of Chow et al. (2008) referred to above. In this study of the Rab-A2 family memb ...
Carey_AminoAcids_Pep..
... These 20 amino acids are the building blocks of proteins. All are a-amino acids. They differ in respect to the group attached to the a carbon. ...
... These 20 amino acids are the building blocks of proteins. All are a-amino acids. They differ in respect to the group attached to the a carbon. ...
Metazoan Remaining Genes for Essential Amino Acid Biosynthesis
... are often capable of working in the reverse reactions and at least some of the remaining enzymes might be used in the degradation steps for their respective amino acid. Thus, the selective pressure relaxation caused by the loss of pathway partners might not be enough to modify these proteins conside ...
... are often capable of working in the reverse reactions and at least some of the remaining enzymes might be used in the degradation steps for their respective amino acid. Thus, the selective pressure relaxation caused by the loss of pathway partners might not be enough to modify these proteins conside ...
Protein structure prediction

Protein structure prediction is the prediction of the three-dimensional structure of a protein from its amino acid sequence — that is, the prediction of its folding and its secondary, tertiary, and quaternary structure from its primary structure. Structure prediction is fundamentally different from the inverse problem of protein design. Protein structure prediction is one of the most important goals pursued by bioinformatics and theoretical chemistry; it is highly important in medicine (for example, in drug design) and biotechnology (for example, in the design of novel enzymes). Every two years, the performance of current methods is assessed in the CASP experiment (Critical Assessment of Techniques for Protein Structure Prediction). A continuous evaluation of protein structure prediction web servers is performed by the community project CAMEO3D.