* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Document 8885981
Survey
Document related concepts
Phosphorylation wikipedia , lookup
Signal transduction wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Magnesium transporter wikipedia , lookup
List of types of proteins wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Protein domain wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein structure prediction wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein folding wikipedia , lookup
Western blot wikipedia , lookup
Protein purification wikipedia , lookup
Transcript
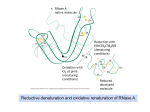
CHEMISTRY SEMINAR “Chaperoning Potential of Disulfide Bond Forming Enzyme” DR. CASSIDY DOBSON DEPARTMENT OF CHEMISTRY & BIOCHEMISTRY ST. CLOUD STATE UNIVERSITY Friday, April 8 12:00 p.m. WSB-122 Abstract: Understanding protein folding is essential in biochemical research. Several diseases are a result of incorrect protein folding and can ultimately lead to the formation of protein aggregates1. Chaperone proteins can assist proper protein folding as a protein matures, but once protein aggregates form generally they cannot be renatured to their individual, correct structure. Disulfide bond forming (DBF) enzyme is a chaperone protein related to the class of Sso7d proteins from the hyperthermophilic bacteria Sulfolobus solfataricus2. These proteins have demonstrated the unique ability to renature proteins from pre-formed protein aggregates. In addition, DBF has been shown to specifically refold proteins with incorrectly made disulfide bonds. Disulfide bonds in protein folding present a more complicated arrangement because of their redox bond formation, which has been show to only be rearranged in the presence of chaperones that themselves contain cysteines residues. DBF is unique to the class of disulfide chaperones in that it does not contain cysteine amino acids, thus presenting a new and different mechanism of disulfide bond rearrangement. We seek to understand the mechanism of action DBF utilizes and apply this mechanism to a variety of different protein aggregates. 1. Dobson, C. (2003) Protein folding and misfolding. Nature Reviews Mol Cell Bio 426, 884-890. 2. Guagliardi, A., Cerchia, L., Camardella, L., Rossi, M., Bartolucci, S. (1994) DBF (Disulfide bond forming) enzyme from the hyperthermophilic archaebacterium Sulfolobus solfataricus behaves like a molecular chaperone. Biocatalysis 11, 181-190. This research provides students the opportunity for undergraduate students to pursue a variety of biochemical endeavors including (but not limited to) molecular biology and cloning, protein expression and purification, biophysical protein characterization and bioinformatics. The techniques employed include a variety of protein purification strategies (IMAC, HIC, Ion-exchange, SEC), as well as enzymatic assays, dynamic light scattering, HPLC, and mass-spectrometry amongst others. Collaborations with other research groups bring the potential for 2D-NMR structural studies, X-ray crystallographic studies, cell culture/physiological studies as well as drug-design and materials science/polymer development.