* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download ppt - The Marko Lab
Public health genomics wikipedia , lookup
Oncogenomics wikipedia , lookup
Non-coding DNA wikipedia , lookup
Genetic testing wikipedia , lookup
Genome evolution wikipedia , lookup
Genetic code wikipedia , lookup
Dual inheritance theory wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Adaptive evolution in the human genome wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Group selection wikipedia , lookup
Designer baby wikipedia , lookup
Human genetic variation wikipedia , lookup
Genetic engineering wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Genome (book) wikipedia , lookup
Frameshift mutation wikipedia , lookup
Gene expression programming wikipedia , lookup
History of genetic engineering wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Point mutation wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Koinophilia wikipedia , lookup
Genetic drift wikipedia , lookup
Population genetics wikipedia , lookup
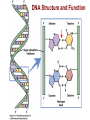
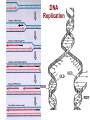
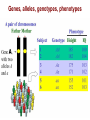
Lecture 3 - Concepts of Marine Ecology and Evolution II 1) Review: Forces of evolution, DNA structure and function, genes, alleles, genotype, phenotype 2) Fitness: viability and fertility 3) Detecting evolution: HW Equilibrium Principle -Calculating allele frequencies, predicting genotypes 4) Detecting selection: LAP evolution in mussels What is Evolution? DNA Structure and Function DNA Replication DNA to Protein: Genotype to Phenotype transcription translation Genes, alleles, genotypes, phenotypes Forces of Evolution 1) Natural selection 2) Gene flow 3) Genetic drift 4) Mutation 5) Non-random mating Darwin’s Theory of Natural Selection Observation 1: Observation 2: Observation 3: Fitness 1) Differential survival: 2) Differential reproductive success: Example A1A1 0.90 3.0 A1A2 0.85 3.0 A2A2 0.80 5.0 Absolute fitness W 2.7 Relative fitness w 0.68 2.55 0.64 4.0 1.0 Prob. of survival # of zygotes Deterministic process = outcome is determined by prior states; predictable outcome Gene Flow (Migration) Mixing of genotypes btn pops Evolutionary consequences of migration Pop. 1, high Pop. 2, high A2 Random migrants (prop to freqs) Genetic Drift A consequence of random mating Y = 0.4 R = 0.2 O = 0.4 N = 100 Y = 0.399 R = 0.188 O = 0.413 N=5 2Y, 2R, 1O Y = 0.4 R = 0.4 O = 0.2 N=5 2Y, 3R, 0O Y = 0.4 R = 0.6 O=0 Genetic Drift Software: AlleleA1 http://faculty.washington.edu/herronjc/SoftwareFolder/AlleleA1.html Genetic Drift If pop size is small enough, for long enough, drift will cause Most Mutations Examples of Adaptive Mutations Malaria resistance: HbS and HbC HbS: heterozygotes resistant HbC: hets: 29% less likely, homoz: 93% less likely Increased bone density (LRP5) Significantly denser and stronger, less skeletal degeneracy Reduced risk of arteriosclerosis (HDLs: Apo-AI-Milano) One copy: HDLs significantly more effective at dissolving arterial plaques HIV resistance (CCR5d32) One copy: AIDs does not develop Two copies: completely resistant to HIV How Often Are Mutations Beneficial? 1 in 150 mutations beneficial 1% fitness advantage Detecting Evolutionary Change G. H. Hardy and W. Weinberg (1908) The HW Equilibrium Principle If no “disturbing influences” act on a genetic locus, allele freqs will remain constant “Disturbing influences” 1) Natural selection 2) Gene flow 3) Genetic drift 4) Mutation 5) Non-random mating Basic Skill: Calculation of allele frequencies Sample of 100 individuals: Genotype AA Aa aa # each allele 2(36) = 72 48 = 48 (of each) 2(16) = 32 N 36 48 16 f(A) = p = 72 + 48 = 120 total number of A alleles = 120/ 200 = 0.6 f(a) = q = 32 + 48 = 80 = 80/ 200 = 0.4 NOTE: 0. 4 + 0.6 = 1 freq of A allele total number of a alleles freq of a allele Predicting Genotype Frequencies What’s the prob 2 gametes meet to form a zygote with a particular genotype? f(A) = p = 0.6 and f(a) = q = 0.4 Possible Genotypes AA Aa aa Predicting Genotype Frequencies What’s the prob 2 gametes meet to form a zygote with a particular genotype? f(A) = p = 0.6 and f(a) = q = 0.4 Converting to Genotype Numbers What’s the prob 2 gametes meet to form a zygote with a particular genotype? f(A) = p = 0.6 and f(a) = q = 0.4 Expected Genotype Frequencies AA = 0.36 Expected #s in a sample of 100: AA = 36 aa = 0.16 aa = 16 Aa = 0.48 Aa = 48 Hardy-Weinberg Equilibrium Principle If no evolutionary forces, we can expect: Generation 1 AA = 36 Aa = 48 aa = 16 p = 0.6 q = 0.4 HWE Expected (84): AA = 42.86 Aa = 34.28 aa = 6.856 p2 = 0.5102 2pq = 0.4082 q2 = 0.0816 Observed (84): AA = 36 Aa = 48 Selection aa = 0 p = 0.7143 q = 0.2857 Generation 2 AA = 51.02 Aa = 40.82 aa = 8.16 p = 0.7143 q = 0.2857 HWE Expected (100): AA = 51.12 Aa = 40.76 aa = 8.12 p2 = 0.5112 2pq = 0.4076 q2 = 0.0812 Observed (100): AA = 51 Aa = 41 No Selection aa = 8 p = 0.7150 q = 0.2850 Are the 5 HWE assumptions ever met? Loci with alleles whose phenotypes have no + or – fitness effects: neutral polymorphisms e.g. blood cell-surface antigens Race and Sanger (1975) – MN genotypes in London MM MN NN Observed 363 634 282 Expected (HWE) 361.54 636.93 280.53 Is HWE for MN blood groups reasonable? -London: large population (12M) -Immigration: low relative to pop size -Heritable mutations are too rare… …to change frequencies -No selective advantage for either allele -Mating is random with respect to blood type 15 - 35 days High dispersal potential 10s to 100s of km 75 85 90 100 Gulf of Maine Long Island Sound 94 98 96 Leucine aminopeptidase (LAP) is an exopeptidase that catalyzes the hydrolysis of amino acid residues from the amino terminus of polypeptide chains. LAPs are widely distributed, ubiquitous in nature, and are of critical biological importance because of their role in protein degradation (Burley et al. 1990). Smaller HW deviation Larger 94 98 96 Gulf of Maine Forces of Evolution 1) Natural selection 2) Gene flow 3) Genetic drift 4) Mutation 5) Non-random mating