* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Media:RuthNov07pres
Survey
Document related concepts
Genetic engineering wikipedia , lookup
Genomic imprinting wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene therapy wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Community fingerprinting wikipedia , lookup
Gene expression wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Metabolic network modelling wikipedia , lookup
Expression vector wikipedia , lookup
Gene desert wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Gene nomenclature wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Transcript
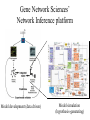
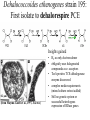
from Newman & Banfield, Science, 2002 Types of models for systems biology From Price & Shmulevich, 2007, Curr Opinion Biotech • Biochemical reaction network Modeling Syntrophic growth of Desulfovibrio & Methanococcus • Selected known pathways and generated two-organism model • 170 reactions; 147 compounds in stoich matrix • Fluxanalyzer program (run via MatLab) • Good predictions of behavior of pure cultures and relative growth rates of orgs in co-culture • In silico & real knockout mutants suggested that interspecies H2-transfer essential and formatetransfer not. From Price & Shmulevich, 2007, Curr Opinion Biotech The GNS framework: a combined approach Genome mining Gene Network Sciences’ Network Inference platform Aksenov et al., 2005 Model development (data driven) Model simulation (hypothesis-generating) GNS framework applied to cancer drug discovery Iterative experiments to Refine models Perturbation (e.g. drug type and level) Cell response (gene regulation) Modified phenotype (e.g. reduced cancer cell division) “heterologous” datasets Network inference engine ID’s genes that are biomarkers for cancer and/or targets for drugs Nodes and edges (interactions) in an inference model Key tools: Bayes theorem Dehalococcoides ethenogenes strain 195: First isolate to dehalorespire PCE Insights gained 0.2 μm (from Maymo-Gatell et al., 1997, Science) • H2 as only electron-donor • obligately uses halogenated compounds as e- acceptors • TceA protein: TCE-dehalogenase enzyme discovered • complex media requirements (mixed culture extract added) • Still no genetic system or successful heterologous expression of RDase genes Highlights from the genome of D. ethenogenes • • 1.5 Mb in size (streamlined) Annotation suggests: – Up to 19 Reductive Dehalogenases (RDases) – 5 Hydrogenases – Vitamin B12 salvage pathways – Other oxidoreductases (including “formate dehydrogenase”) that might be directly involved in dehalorespiration – Evidence of extensive horizontal gene transfer – Lesions in key intermediary pathways (TCA cycle; amino acid biosynth) – Lots of unknown topology (even around RDases (from Seshadri et al., Science 2005) Ethene S-LAYER H C=C H H Cl H Cl HCl PERIPLASM ?? Tetrachloroethene HCO2– CO2 C =C H2 Cl Cl 2H++2e- 2H+ H2 H+ H+ RD H+ Hup Mod Fdh ATP Nuo CYTOPLASM NADH? F420? H2? CO+H2O N2+ 8H++ 8e-+16 ATP e- Ferredoxin 2Fe-Fs Desulforedoxin Glutaredoxin Rubredoxin Flavodoxin Thioredoxin CODH Nif H2 2H+ Hym H2 2H+ Hyc 2H+ Ech H2 CO2+H2 Cl 2NH3 +H2 +16 ADP+16 Pi Cl C=C 2H+ Vhu Cl Cl Signal? Redox potential PMF Response regulator = Fe H2ase large subunit = Molybdopterin-containing subunit = NiFe H2ase large subunit H2 H+ NADNADH FADFADH His kinase sensor RD RD anchoring protein ADP + Pi Some key questions the gene network modeling will address: • What networks of RDases emerge in cultures grown on different substrates? Are there specific transcriptional regulators with expression tied to individual or groups of RDases? • Are individual RDases co-regulated with other elements of the proposed electron transport chain (e.g Hup)? • Which genes are co-regulated with highly-expressed genes of unknown function: “Fdh” and DET00754/755 – each of which were found in all DHC cultures in high abundance. • Which gene networks correlate with the presence of other community members? Does this provide any insight regarding the nutritional benefit to DHC of mixed culture growth? • Which, if any, networks are sensitive to hydrogen concentrations? • How do candidate bioindicators (highlighted in Preliminary Results) correlate with respiration rate over a wider range of growth conditions? • Which biomarkers are indicative of DHC stress DoD project (5/07-12/09): DET mixed cult focused • Overall objective is to develop a whole-cell model of gene networks in DHC that relates growth conditions to gene expression levels and, in turn, relates these levels to dehalorespiration rates. • Approach framework will be to quantitatively monitor genome-wide RNA and protein levels in a model DHC strain (D. ethenogenes strain 195 - DET) growing in mixed-culture conditions in pseudo-steady-state reactors and to utilize systems biology algorithms of network inference to compile the data into a model NSF (9/07 – 8/10):KB-1 focused • The overall objective of the proposed work is to understand how two well-studied DHC cultures respond to environmental conditions and how DHC gene expression can be monitored to inform enhanced bioremediation and forecast modeling efforts at contaminated field sites. The three main objectives (Phases) are: – Objective/Phase 1: Develop in-depth models of gene networks for two wellstudied DHC growing in mixed culture conditions. Here, we aim to determine key gene networks in the DHC that correlate with the type and rate of dechlorination and that indicate how these organisms respond to stressors. – Objective/Phase 2. Test model predictions for one of the DHC models (the bioaugmentation culture KB1) under various field conditions. – Objective/Phase 3. Determine robust quantitative chloroethene dehalorespiration bioindicators and develop qRTPCR and RNA-biosensor assays for them. Perturbations and data types • Perturbations/intervention s (n=30-50 initially) • Variations in – Type and loading rate of chlorinated compounds – Type and loading rate of electron donor – Culture density – Stressors (Oxygen, pH, chloroform). • Datasets to be collected – Omics (microarray; proteomics) – Metabolites (organic acids; H2) – Populations of DHC & other orgs (qPCR) – General activity of pop’ns (qRTPCR) – Chlorinated substrates & products – Dechlorination rates (phenotype of interest)