* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 生物計算
Survey
Document related concepts
Genetic code wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
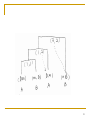
Metagenomics wikipedia , lookup
Oncogenomics wikipedia , lookup
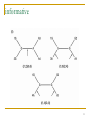
Nucleic acid analogue wikipedia , lookup
Microevolution wikipedia , lookup
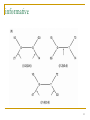
Gene expression programming wikipedia , lookup
Frameshift mutation wikipedia , lookup
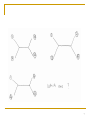
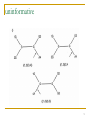
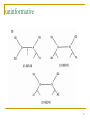
Quantitative comparative linguistics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Helitron (biology) wikipedia , lookup
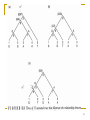
Point mutation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Transcript
Chapter 5 Character–Based Methods of Phylogenetics 暨南大學資訊工程學系 黃光璿 (HUANG, Guan-Shieng) 2004/04/05 1 5.1 Parsimony Mutations are exceedingly rare events. The most unlikely events a model invokes, the less likely the model is to be correct. The fewest number of mutations to explain a state is the most likely to be correct. 2 Ockham's Razor the philosophic rule states that entities should not be multiplied unnecessarily 3 4 5 5.1.1 Informative and Uninformative Sites 6 7 5.1.1 Informative and Uninformative Sites informative sites have information to construct a tree uninformative sites have no information in the sense of parsimony principle. 8 uninformative 9 uninformative 10 informative 11 informative 12 A position to be informative must have at least two different nucleotides each of these nucleotides to present at least twice. 13 informative sites synapomorphy: support the internal branches (true) homoplasy: acquired as a result of parallel evolution of convergence (false) 眼睛:humans, flies, mollusks (軟體動物) 14 5.1.2 Unweighted Parsimony Every possible tree is considered individually for each informative site. The tree with the minimum overall costs are reported. 15 16 There are several problems: The number of alternative unrooted trees increases dramatically. Calculating the number of substitutions invoked by each alternative tree is difficult. 17 The second problem can be solved by intersection: if the intersection of the two sets of its children is not empty union: if it is empty. The number of unions is the minimum number of substitutions. For uninformative site, it is the number of different nucleotides minus one. 18 /* the uth position in the kth sequence */ 19 5.1.4 Weighted Parsimony Not all mutations are equivalent Some sequences (e.g., non-coding seq.) are more prone to indel than others. Functional importance differs from gene to gene. Subtle substitution biases usually vary between genes and between species. Weights (scoring matrices) can be added to reflect these differences. 20 21 22 23 24 Calculating the optimal costs 25 Finding the internal nodes 26 5.2 Inferred Ancestral Sequences Can be derived while constructing the tree. No missing link! 如何取樣本? It may be bias. 27 5.3 Strategies for Faster Searches The number of different phylogenetic tree grows enormously. 10 sequences 2M for exhaustive search 28 參考資料及圖片出處 1. 2. 3. Fundamental Concepts of Bioinformatics Dan E. Krane and Michael L. Raymer, Benjamin/Cummings, 2003. Biological Sequence Analysis – Probabilistic models of proteins and nucleic acids R. Durbin, S. Eddy, A. Krogh, G. Mitchison, Cambridge University Press, 1998. Biology, by Sylvia S. Mader, 8th edition, McGraw-Hill, 2003. 52