* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download DNA Recombination
Oncogenomics wikipedia , lookup
DNA barcoding wikipedia , lookup
DNA profiling wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
Metagenomics wikipedia , lookup
Genetic engineering wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Mitochondrial DNA wikipedia , lookup
SNP genotyping wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Designer baby wikipedia , lookup
Primary transcript wikipedia , lookup
Human genome wikipedia , lookup
Cancer epigenetics wikipedia , lookup
DNA polymerase wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Genome evolution wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
DNA vaccination wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Transposable element wikipedia , lookup
Molecular cloning wikipedia , lookup
Epigenomics wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Point mutation wikipedia , lookup
Microsatellite wikipedia , lookup
Genomic library wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Microevolution wikipedia , lookup
DNA supercoil wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
History of genetic engineering wikipedia , lookup
Non-coding DNA wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genome editing wikipedia , lookup
Homologous recombination wikipedia , lookup
Holliday junction wikipedia , lookup
DNA Recombination
•
•
•
•
Roles
Types
Homologous recombination in E.coli
Transposable elements
Biological Roles for Recombination
1. Generating new gene/allele combinations
(crossing over during meiosis)
2. Generating new genes (e.g., Immunoglobulin rearrangement)
3. Integration of a specific DNA element
4. DNA repair
Practical Uses of Recombination
1. Used to map genes on chromosomes
(recombination frequency proportional
to distance between genes)
2. Making transgenic cells and organisms
Map of Chromosome I of
Chlamydomonas reinhardtii
cM = centiMorgan; unit of recombination frequency
1 cM = 1% recombination frequency
Chlamydomonas Genetics Center
Types of Recombination
1. Homologous - occurs between sequences
that are nearly identical (e.g., during
meiosis)
2. Site-Specific - occurs between sequences
with a limited stretch of similarity; involves
specific sites
3. Transposition – DNA element moves from
one site to another, usually little sequence
similarity involved
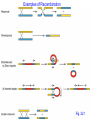
Examples of Recombination
Fig. 22.1
Holliday Model
R. Holliday (1964)
- Holliday Junctions
form during
recombination
- HJs can be resolved
2 ways, only one
produces true
recombinant
molecules
Fig. 22.2
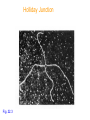
patch
EM of a Holliday Junction w/a few melted
base pairs around junction
Fig. 22.3
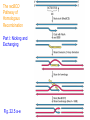
The recBCD
Pathway of
Homologous
Recombination
Part I: Nicking and
Exchanging
Fig. 22.5 a-e
recBCD Pathway of Homologous
Recombination
Part I: Nicking and Exchanging
1.
2.
3.
4.
5.
A nick is created in one strand by recBCD at a Chi
sequence (GCTGGTGG), found every 5000 bp.
Unwinding of DNA containing Chi sequence by
recBCD allows binding of SSB and recA.
recA promotes strand invasion into homologous
DNA, displacing one strand.
The displaced strand base-pairs with the single
strand left behind on the other chromosome.
The displaced and now paired strand is nicked
(by recBCD?) to complete strand exchange.
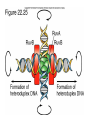
recBCD Pathway
of Homologous
Recombination
Part II: Branch
Migration and
Resolution
Fig. 22.5 f-h
recBCD Pathway of Homologous Rec.
Part II: Branch Migration and Resolution
1. Nicks are sealed Holliday Junction
2. Branch migration (ruvA + ruvB)
3. Resolution of Holliday Junction (ruvC)
RecBCD : A complex enzyme
• RecBCD has:
1. Endonuclease subunits (recBCD) that cut
one DNA strand close to Chi sequence.
2. DNA helicase activity (recBC subunit) and
3. DNA-dependent ATPase activity
– unwinds DNA to generate SS regions
Activity 2 and 3 linked
RecA
• 38 kDa protein that polymerizes onto SS DNA 5’-3’
• Catalyzes strand exchange, also an ATPase
• Also binds DS DNA, but not as strongly as SS
Fig. 22.6
RecA Function Dissected
• 3 steps of strand exchange:
1. Pre-synapsis: recA coats single stranded
DNA (accelerated by SSB, get more relaxed
structure, Fig. 22.8)
2. Synapsis: alignment of complementary
sequences in SS and DS DNA (paranemic
or side-by-side structure)
3. Post-synapsis or strand-exchange: SS DNA
replaces the same strand in the duplex to
form a new DS DNA (requires ATP
hydrolysis)
RuvA and RuvB
• DNA helicase that catalyzes branch migration
• RuvA tetramer binds to HJ (each DNA
helix between subunits)
• RuvB is a hexamer ring, has helicase & ATPase
activity
• 2 copies of ruvB bind at the HJ (to ruvA and 2 of
the DNA helices)
• Branch migration is in the direction of recA
mediated strand-exchange
http://www.sdsc.edu/journals/mbb/ruva.html
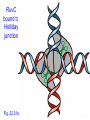
RuvC : resolvase
• Endonuclease that cuts 2 strands of HJ
• Binds to HJ as a dimer
• Consensus sequence: (A/T)TT (G/C)
- occurs frequently in E. coli genome
- branch migration needed to reach
consensus sequence!
RuvC
bound to
Holliday
junction
Fig. 22.31a
Transposable Elements
(Transposons)
• DNA elements capable of moving ("transposing")
around the genome
• Discovered by Barbara McClintock, largely from
cytogenetic studies in maize, but since found
in most organisms
• She was studying "variegation" or sectoring in
leaves and seeds
• She liked to call them "controlling elements“
because they effected gene expression in
myriad ways
Mutant Kernel Phenotypes
•
•
Pigmentation mutants
– affect anthocyanin pathway
– elements jump in/out of transcription
factor genes (C or R)
– sectoring phenotype - somatic mutations
– whole kernel effected - germ line
mutation
Starch synthesis mutants
- stain starch with iodine, see sectoring in
endosperm
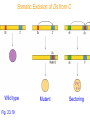
Some maize phenotypes caused by transposable
elements excising in somatic tissues.
Start with mutant kernels defective in starch synthesis (endosperm
phenotypes) or anthocyanin synthesis (aleurone and pericarp
phenotypes).
Somatic Excision of Ds from C
Wild type
Fig. 23.19
Mutant
Sectoring
Other Characteristics of McClintock's
Elements
• Unstable mutations that revert frequently but often
partially, giving new phenotypes.
• Some elements (e.g., Ds) correlated with
chromosome breaks.
• Elements often move during meiosis and
mitosis.
• Element movement accelerated by genome
damage.
Molecular analysis of transposons
• Transposons isolated by first cloning a gene that
they invaded. A number have been cloned this way,
via "Transposon trapping“.
• Some common molecular features:
– Exist as multiple copies in the genome
– Insertion site of element does not have extensive
homology to the transposon
– Termini are an inverted repeat
– Encode “transposases” that promote movement
– A short, direct repeat of genomic DNA often
flanks the transposon : “Footprint”
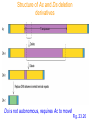
Ac and Ds
•
•
•
•
Ds is derived from Ac by internal deletions
Ds is not autonomous, requires Ac to move
Element termini are an imperfect IR
Ac encodes a protein that promotes
movement - Transposase
• Transposase excises element at IR, and also
cuts the target
Structure of Ac and Ds deletion
derivatives
Ds is not autonomous, requires Ac to move!
Fig. 23.20
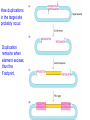
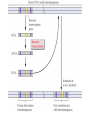
How duplications
in the target site
probably occur.
Duplication
remains when
element excises,
thus the
Footprint.
Mutator (A Retrotransposon)
• Discovered in maize; differs significantly from
Ac by structure and transposing mechanism
• Autonomous and non-autonomous versions;
many copies per cell
• contains a long terminal IR (~200 bp)
• transposes via a replicative mechanism,
instead of a gain/loss mechanism
• A “retrotransposon”
– Similarities with retroviruses
– move via an RNA intermediate
– encode a reverse transcriptase activity
Fig. 7.34 in Buchanan et al.
Control of Transposons
• Autoregulation: Some transposases are
transcriptional repressors of their own
promoter(s)
• e.g., TpnA of the Spm element
• Transcriptional silencing: mechanism not well
understood but important; correlates with
methylation of the promoter
Biological Significance of
Transposons
• They provide a means for genomic change
and variation, particularly in response to
stress (McClintock’s "stress" hypothesis)
(1983 Nobel lecture, Science 226:792)
• or just "selfish DNA"?
• No known examples of an element playing a
normal role in development.