* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Proteomics techniques used to identify proteins
Endomembrane system wikipedia , lookup
Biochemistry wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Gene regulatory network wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Gene expression wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Protein domain wikipedia , lookup
Homology modeling wikipedia , lookup
Gel electrophoresis wikipedia , lookup
Magnesium transporter wikipedia , lookup
Protein folding wikipedia , lookup
Metalloprotein wikipedia , lookup
Expression vector wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
Interactome wikipedia , lookup
Protein moonlighting wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein adsorption wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Western blot wikipedia , lookup
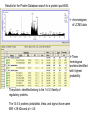
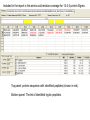
Identification of regulatory proteins from human cells using 2D-GE and LC-MS/MS Victor Paromov Christian Muenyi William L. Stone Proteomics techniques used to identify proteins • Obtain cell lysates • Run two-dimensional gel electrophoresis (2D-GE) for each sample (in triplicates) • Compare the gels and identify over-expressed and/or under-expressed proteins compared to protein spots from control treatment • Identify those proteins (for each spot) by: – Cutting out the gel spot and trypsinizing the protein – Run LS/MS/MS on LTQ XL – Identify proteins by matching the tryptic peptides to theoretical fragments (Protein Database search) 2DGE Example comparing Vehicle- vs CEES-treated human keratinocytes Vehicle 2.5 mM CEES S #1 S #2 Proteomics Study of CEES toxicity in human keratinocytes: EpiDerm tissues were exposed to vehicle or 2.5 mM CEES for 18 h. Cell lysates were separated by 2D gel electrophoresis, Coomassie blue-stained and photographed (three gels per sample). Average differences in protein expression were quantified with Dymension-2 Software. The proteins differentially expressed after CEES exposure are marked with red circles. Protein spots were excised from the gel, destained, trypsinized, and subjected to LC/MS/MS analysis. Results for the Protein Database search for a protein spot #543. ← chromatogram of LC/MS data ←Three homologous proteins identified with highest probability. The protein identified belong to the 14-3-3 family of regulatory proteins. The 14-3-3 proteins (zeta/delta, theta, and sigma) have same MW = 28 kDa and pI = 4.5. Included in the report is the amino acid residue coverage for 14-3-3 protein Sigma. Top panel: protein sequence with identified peptides (shown in red). Bottom panel: The list of identified tryptic peptides. MS/MS fragment ion matches for a peptide fragment of 14-3-3 protein Sigma (YLAEVATGDDK) The list of B and Y ions as they match against the experimental data (matched B ions – red, matched Y ions – blue). The mass errors of the measurement. MS/MS data of fragment ions that matched theoretical data of database Conclusion: LC/MS/MS analysis and UniProt database search with “rigorous” filtering of the search results allow identification of three 14-3-3 regulatory proteins (zeta, theta, and sigma).