* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Breanna Perreault D145 Presentation 2/23/17 Background
Extrachromosomal DNA wikipedia , lookup
Genetic engineering wikipedia , lookup
Metagenomics wikipedia , lookup
Human genome wikipedia , lookup
Gene expression programming wikipedia , lookup
History of RNA biology wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Genome evolution wikipedia , lookup
RNA silencing wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
X-inactivation wikipedia , lookup
Oncogenomics wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Non-coding RNA wikipedia , lookup
Point mutation wikipedia , lookup
Non-coding DNA wikipedia , lookup
Gene expression profiling wikipedia , lookup
History of genetic engineering wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Helitron (biology) wikipedia , lookup
Designer baby wikipedia , lookup
Microevolution wikipedia , lookup
Epitranscriptome wikipedia , lookup
Genomic imprinting wikipedia , lookup
Epigenetic clock wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Primary transcript wikipedia , lookup
Histone acetyltransferase wikipedia , lookup
DNA methylation wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Epigenetics of depression wikipedia , lookup
Transgenerational epigenetic inheritance wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Behavioral epigenetics wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Epigenetics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Epigenomics wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
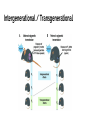
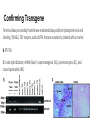
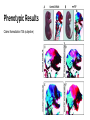
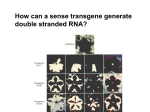
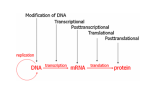
Breanna Perreault D145 Presentation 2/23/17 Background: Epigenetics Why are we studying? - Traditional Genetics Insufficient Explanation What is Epigenetics? - A change in phenotype without a change in genotype; alteration of gene expression without alteration of endogenous DNA code Epigenetics Vocabulary If you have a smartphone, I recommend taking a picture of this slide now :) Histone Methylation: Methylation on Histones DNA Methylation: Direct Methylation on DNA, especially on CpGs CpGs: Consecutive C and G nucleotides, sequence that can be directly methylated CpG Islands: Areas with very high CG content, can help regulate transcription Intergenerational: All “existent” at time of subject’s examination Transgenerational: All following generations yet to be created Knowns Before Entering The Experiment Father’s experiences seems to have an effect on progeny -Past Studies Methylated CpGs not responsible for implied heritability in some studies - CpG methylation heritable Overview: Variant Histone (not DNA) Methylation and Sperm RNA linked to altered transgenerational gene expression H3K4me2 (Dimethylated Histones on transcription factor binding regions)on developmental genes reduced by expression of KDM1A histone lysine 4 demethylase Paternal Epigenetic Inheritance Effects in development and fitness lasted two generations, including those without mutant genotype (heritable) Intergenerational / Transgenerational Background: Paternal Inheritance This study focused on Spermatogenesis from Primordial Germ Cells (or PGCs) - Rapid cell division - Highly compacted nucleus- protamine - Low histone levels Promoter regions of housekeeping genes : histone variant H3.3, H3K4 di- and trimethylation. Developmental regulatory genes: H3.3, H3.1 &H3.2 variants & trimethylation of H3K27 Creation of Transgenic Animals Microinjection into Embryos P EF1a - Translation elongation factor EF1A (gonad specific, germ not somatic) IRES - internal ribosome entry site EGFP - Green Flourescent Protein Confirming Transgene Terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL). TdT enzyme, adds dUTPs that are secondarily labeled with a marker. B RT PCR C In situ hybridization; mRNA (blue) in spermatogonia (SG), spermatocytes (SC), and round spermatids (RS) Experimental Breeding Crosses Phenotypic Results Phenotypic Results Claims Normalization TG4 (subjective) Mortality Results Proposed Mechanism of Variable Deformities Mortalities and abnormalities non- mendelian -KDM1A is active in spermatogenesis; diploid cells. -Cellular bridges from incomplete mitotic and meiotic divisions; transgene in 50% of spermatids but all haploid spermatids affected Histone Localization Analysis Which genes are affected? 1. DTT treatment - remove protamine 2. Micrococcal Nuclease digestion 3. Chromatin immunoprecipitation (ChIP) with H3K4me2specific antibodies 4. ChIP-seq ( DNA to protein, next gen, massively parallel) ChIP-seq Location of Demethylation (Chip Seq Results) KDM1A to transcription start sites with H3.3 and H3K4me2 Therefore with high CpG content -Gene ontology H3K4me2 involved in morphogenesis, patterning, vasculature development, cartilage development, and catabolic and metabolic processes KDM1A preferentially alters offspring development. Gene Ontology Pdpk1 = embryogenesis, growth, and nervous system development E2F6 = skeletal defects Gsc = craniofacial abnormalities F3 Progeny has reduced H3K4me2 Proposed Mechanism: TSS (transcription start site )regions with reduced H3K4me2 levels are strongly enriched for H3.3 High levels of H3.3 = high nucleosome turnover at CGIs in spermatids KDM1A cannot maintain H3K4me2 state during nucleosome turnover in spermatids of TG3 males. F3 Progeny has reduced H3K4me2 Similar demethylation methylation to start, Mature TG changes MATURE Spermatozoa PRE-Spermatazoa Any other way Non-TG offspring got phenotypic aberrations? Considering methylation mechanism of phenotypic aberrations in Non-TG offspring - Target selection: CpGs differentially methylated in TG3 sperm & C57BL/6 controls - Quantitative MassARRAY (with bisulfite) - RBBS (Reduced representation bisulfite sequencing) Result: Not methylation MassARRAY iPLEX assay Extension primer RBBS (Reduced representation bisulfite sequencing) RNA products of Overexpression of KDM1A Late Affymetrix GeneChip ST2.0 Array for RNA content Affymetrix gene chip Early Overlapping Mutant Genotypes Findings Critique Purpose of study is often explained as environmental, but methods not environmental. Distinction between RNA or altered Histones as reason for epigenetic inheritance Use of human gene to create the experimental variable in the mouse Phenotypic analysis of the transgenic pups not quantitative Borrowed Data Bibliography & Further Reading This article: Keith Siklenka1,*, Serap Erkek2,3,*,†, Maren Godmann4,‡, Romain Lambrot4, Serge McGraw5, Christine Lafleur4, Tamara Cohen4, Jianguo Xia4,9, Matthew Suderman7, Michael Hallett8, Jacquetta Trasler5,6, Antoine H. F. M. Peters2,3,*,§, Sarah Kimmins1,4,*,§1Dep, Keith Siklenka, Serap Erkek, Maren Godmann, Romain Lambrot, Serge McGraw, Christine Lafleur, Tamara Cohen, Jianguo Xia, Matthew Suderman, Michael Hallett, Jacquetta Trasler, Antoine H. F. M. Peters, Sarah Kimmins, and Published Online08 Oct 2015. "Disruption of Histone Methylation in Developing Sperm Impairs Offspring Health Transgenerationally." Disruption of Histone Methylation in Developing Sperm Impairs Offspring Health Transgenerationally | Science. N.p., n.d. Web. 23 Feb. 2017. http://science.sciencemag.org/content/350/6261/aab2006/tab-pdf Article on the histone analysis method: Caglayan, A. O., Comu, S., Baranoski, J. F., Parman, Y., Kaymakçalan, H., Akgumus, G. T., et al. (2015). NGLY1 mutation causes neuromotor impairment, intellectual disability, and neuropathy. Eur J Med Genet, 58(1), 39-43. http://www.nature.com/nprot/journal/v8/n12/full/nprot.2013.145.html Article on an interesting paternal epigentic phenomenon B. G. Dias, K. J. Ressler, Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat. Neurosci. 17, 89–96 (2014) http://www.nature.com/neuro/journal/v17/n1/full/nn.3594.html Hay Preguntas?