* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Homologous Recombination DNA break repair by homologous
Mitochondrial DNA wikipedia , lookup
DNA profiling wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
X-inactivation wikipedia , lookup
Genome (book) wikipedia , lookup
Human genome wikipedia , lookup
Neocentromere wikipedia , lookup
Primary transcript wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Genetic engineering wikipedia , lookup
DNA polymerase wikipedia , lookup
Genome evolution wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Cancer epigenetics wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Genealogical DNA test wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
Transposable element wikipedia , lookup
SNP genotyping wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Designer baby wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Epigenomics wikipedia , lookup
DNA vaccination wikipedia , lookup
Point mutation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Genomic library wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Non-coding DNA wikipedia , lookup
Molecular cloning wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Microevolution wikipedia , lookup
DNA supercoil wikipedia , lookup
History of genetic engineering wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genome editing wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Holliday junction wikipedia , lookup
Homologous recombination wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
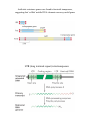
Homologous Recombination • repair of dsDNA damage • recombination between homologous chromosomes STEP 1 create ssDNA with free 3’OH ssW1 dsC2-W2 STEP 2 find homology by strand exchange: STEP 3 extend region of strand exchange beyond initial homology STEP 4 resolve junction of dsDNAs to reestablish 2 separate chromosomes DNA break repair by homologous recombination This requires specialized factors: a protein helps the ssDNA region to find the homologous dsDNA in order to trade base-pairing. ssW2 dsC2-W1 STEP 1: Create ssDNA with free 3’ OH Eukaryotes typically load a 5’-3’ exonuclease at a dsDNA break. Also possible to nick DNA then load a helicase: 5’ 3’ 3’ 5’ 3’ 5’ 3’ 5’ In E. coli, homologous recombination is induced by RecBCD RecB and RecD are helicases with opposite polarity. They load as a complex with each other and RecC at a break. Rec B is also a nuclease; it cuts both single strands generated by the helicases UNTIL it encounters (running in the right polarity) the ‘chi’ site, at which point it leaves the strand with the free 3’ OH alone and continues to degrade the other strand. STEP 2: Strand exchange to find homology E. coli uses RecA The ssDNA in a RecA filament threads past dsDNA, with base flipping from the dsDNA acting to sample homology with the ssDNA. Binding to RecA induces underwinding of the DNA, which encourages bases to flip back and forth between the two possible partner strands WITHOUT additional input of energy. Model for DNA strand exchange mediated by RecA. A three-strand reaction is shown. (a) RecA protein forms a filament on the single-stranded DNA. (b) A homologous duplex incorporates into this complex. (c) One of the strands in the duplex is transferred to the single strand originally bound in the filament. The other strand of the duplex is displaced. Important features of RecA: • A monomer binds ~3 nt or bp • Cooperative filament assembly 5’-3’ • Prefers to form filament on ssDNA, but once formed, the filament will take up dsDNA at a second site • Filament has 18.6 bp DNA/turn: bp are de-stabilized and can rapidly exchange between two bound DNAs • Bound ATP increases RecA DNA affinity, ATP hydrolysis decreases affinity for DNA Human cells have the RecA-like protein Rad51; Rad51 needs extra help STEP 2 Products Each ssDNA strand exchanged generates a Holliday junction. Several series of steps are possible, so ONLY consider the model junction below. **Stable strand exchange by RecA requires >50 bp of PERFECT homology C1 W1 W2 C2 3’ 5’ 5’ 3’ 3’ 3’ STEP 3: Extend region of strand exchange “Branch Migration” of the Holliday junction C1 W1 W2 C2 3’ 5’ 5’ 3’ C1 W1 W2 C2 heteroduplex C1 W1 OR heteroduplex W2 C2 The Holliday junction is held in square-planar configuration by a sandwiching octamer of RuvA C1 W1 C1 W2 parental W2 C2 heteroduplex W2-C1 heteroduplex W1 C2 parental W2 C2 C1 W1 parental C2-W1 heteroduplex RuvA (in green) maintains Holliday junction geometry, recruits RuvB RuvB (in white) is a hexameric helicase; it extends the heteroduplex parental W2 C2 **RuvB is ATP-powered: heteroduplex formation can proceed WITHOUT perfect homology, over long (>1 kb) regions heteroduplex W2-C1 parental C1 W1 C2-W1 heteroduplex Holliday junction resolution: the endonuclease RuvC (E. coli) It must nick BOTH Crick strands OR BOTH Watson strands to separate the two duplex DNAs (different chromosomes) W2-C1 heteroduplex C1 W1 C1 W2 W2 C2 W1 C2 Cut at BOTH thin OR BOTH thick arrows W2 C2 parental C1 W1 parental C2-W1 heteroduplex (eukaryotic equivalents of RuvABC have been purified but their identity is not certain) C1 W2 C1 W1 W2 C2 C1 W1 W2 C2 W1 C2 C1 W1 W1 C2 W2 C2 C1 W2 C1 W1 W2 C2 OR C1 W1 W2 C2 3’ C1 W2 100% chance of some heteroduplex 50% chance of recombinant ends (exchange of chromosome arms) C1 W1 5’ 5’ 3’ W1 C2 W2 C2 Gene conversion can make heterozygous loci homozygous (called loss of heterozygousity or LOH) Products of HR (shown with recombinant ends, but note that central heteroduplex is present even with parental ends): Large-scale genome rearrangments by inappropriate HR RuvC cleaves to give back parental ends RuvC cleaves to give recombinant ends: deletion, inversion and translocation events Homologous recombination with RECOMBINANT ENDS that occurs between duplicated genes (or other duplicated loci) can result in chromosome deletion, inversion and translocation events DELETION INVERSION chromosome 1 chromosome 2 TRANSLOCATION Site-specific recombination Protein-DNA recognition at sites with a specific sequence The two sites ‘synapse’ then all four strands are cut in series to exchange the original ends for recombinant ends. Performed by a tetramer of a site-specific recombinase. The enzyme active site tyrosine forms a covalent protein-DNA intermediate like a topoisomerase, so the recombination reaction is reversible with no need for DNA ligase. Site-specific recombination The only difference between the reactions in (A) and (B) is the relative orientation of the two DNA sites (indicated by arrows) at which a site-specific recombination event occurs. Why bother with sitespecific recombination? A surface contour model of Cre recombinase bound to a recombination intermediate. The protein has been rendered transparent so that the bound DNA is visible. Lambda phage hides in the E. coli chromosome by integration: attB (bacterium): 25 bp attL (left) attP (phage): 240 bp attR (right) Salmonella evades the immune system by changing gene expression: gene on (gene off) Example of a DNA transposon IS = “insertion sequence” for the mode of its discovery Non-replicative (cut-and-paste) transposition. DNA-based transposons have an inverted repeat sequence at their ends, and any DNA between them can be moved. Transposase multimers make a blunt double-stranded cut at the edge of the inverted repeat termini. Transposase also has a second binding site for DNA that is not sequence-specific, which it uses to bind an insertion target site and make a staggered double-stranded cut. Transposase bound to the transposon ends reverses its cleavage reaction to ligate the transposon DNA to the target site ends, but a gap remains on each side of the inserted DNA due to the staggered target site cut. Repair synthesis is required to rejoin the broken donor chromosome and to fill in the target site gaps. Different transposase enzymes make different types of staggered cuts. Depending on order of the next steps, transposition can result in transposon movement or transposon retention at the donor site and insertion elsewhere as well. If transposase nicks the donor site ends rather than cutting both strands at once then donor 3’ ends join target 5’ ends, target 3’ ends prime replication and result in duplication of the transposon. The resulting donor-target fusion is fixed by the activity of a transposon-encoded site-specific recombinase or ‘resolvase’. Importantly, even if the transposon departs from the donor site, the target site direct repeat is left behind (this is mutagenic). Antibiotic resistance genes were found in bacterial transposons, suggesting that ‘selfish’ mobile DNA elements can carry useful genes LTR (long terminal repeat) retrotransposons transposase a virally encoded integrase enzyme pastes the virus into the host chromosome (like a transposase second step). The life cycle of an LTR retrovirus (like HIV) A Non-LTR retrovirus lifecycle (like the LINE elements that constitute 21% of our genome) Additional mutagenesis occurs from homologous recombination between transposable elements DELETION INVERSION chromosome 1 chromosome 2 TRANSLOCATION Some examples of single-gene diseases Manipulating the genome using endogenous DNA repair to perform gene conversion A B C D E F G a b c d e f g A B C D E F G a b c D e f g X ray-induced break ! Homologous Recombination sister chromatid ! DNA replaced with same sequence induced break ! Induce a dsDNA break at the mutation site to be repaired. donor DNA ! Provide a plasmid template for homologous recombination target replaced with donor DNA sequence ! (plasmid without origin is lost) Zinc Finger Protein (ZFP)-Nucleases Site-specific DNA breaks could be used for gene correction or gene disruption DSB + donor DNA? no non-homologous end-joining deletion? yes homologous recombination gene conversion