* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download File - Mrs. LeCompte
DNA vaccination wikipedia , lookup
History of genetic engineering wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Polyadenylation wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Epigenomics wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Frameshift mutation wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
History of RNA biology wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Non-coding RNA wikipedia , lookup
Point mutation wikipedia , lookup
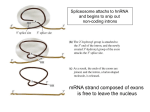
Primary transcript wikipedia , lookup
Messenger RNA wikipedia , lookup
Genetic code wikipedia , lookup
Epitranscriptome wikipedia , lookup
12-5 Notes: Translation Protein Synthesis Original information DNA gets transcribed into mRNA Next Step: to use the mRNA strand to build the polypeptide chain mRNA carries the message as codons = 3 bases at a time o mRNA code is used to determine the actual order of amino acids tRNA’s job is to bring the correct amino acid and place it in the proper position by matching up the codon of mRNA to an anticodon on tRNA tRNA more than 20 kinds in each cell (1 or more for each kind of a.a. in cell) o Wobble Hypothesis = only 40 (not 61) tRNAs are needed in cells because some can pair with more than one codon Helps to explain why the code is degenerate Cloverleaf shape (see diagram below) two attachment sites: o Anticodon = binds to codon on mRNA o 3’ end = attaches to a particular amino acid tRNA Loading Amino Acyl-tRNA Synthetase = enzyme that catalyzes the attachment of an amino acid to its tRNA 20 types of them 2 steps: 1) Activation of the amino acid with ATP: The enzyme’s active site binds the amino acid and ATP ATP is cleaved, releasing 2 phosphates (and energy) and attaches to the amino acid as AMP 2) Attachment of the amino acid to 3’ End (of tRNA) AMP releases bond forms between the a.a. and 3’ end of tRNA amino acid–tRNA complex is released Later, when the tRNA anticodon matches to mRNA codon, the bond breaks between tRNA and the a.a. and the tRNA is released, going back to pick up another a.a. Translation also requires rRNA and tRNA Made in the nucleolus differ structurally and functionally from mRNA Ribosomes made of two subunits: one smaller, one larger 60% rRNA and 40% protein Prokaryotic ribosomes are smaller and have different molecular composition o This is how some antibiotics work attack structure of prokaryotic ribosomes without affecting eukaryotic ribosomes Smaller subunit = binding site for mRNA o 5’ end of mRNA attaches here Larger subunit has binding sites for tRNA Has 3 binding sites: A (amino acid) site, P (polypeptide) site, and E (exit) site PROTEIN SYNTHESIS: Translation (how polypeptides are assembled: mRNA a.a.) 3 stages: 1) Initiation Smaller ribosomal subunit attaches to mRNA near 5’ end o exposes initiator codon o 1st tRNA pairs with initiator codon: usually (5’) – AUG – (3’) Brings modified a.a. = fMet (which may later be removed) Larger ribosomal subunit attaches to smaller subunit o initiator tRNA locked into the P site energy comes from GTP 2) Elongation next complementary mRNA codon is exposed in the A site the next tRNA anticodon and it a.a. are inserted when both spots (A & P) are occupied peptidyl transferase helps form a peptide bond between the two amino acids ribosome slides over one codon and process repeats when the ribosome moves, the tRNA in the P site is moved to the E site, and that tRNA detaches and it exits the ribosome when 5’ end portion of mRNA is free again, another ribosome can attach and build another chain o Polyribosome = a group of ribosomes reading the same mRNA molecule 3) Termination “Stop” codon is near end of mRNA o UAA, UGA, or UAG o NO tRNA’s have the anticodon for these release factor is inserted translation stops and polypeptide chain is released ribosomal subunits separate all parts are reused or degraded After Translation The first few amino acids of a polypeptide act as a signal peptide that indicate whether a protein is for export or not Those polypeptides that are for secretion are then fed into the lumen of the ER, where they are folded and modified by the addition of sugars, phosphates, or lipids o Since prokaryotes do not have ER, this is a limitation on their use in genetic engineering of eukaryotic proteins EUKARYOTIC CHROMOSOMES More than 60% protein Humans have 2 m of DNA packed into a nucleus that is 5 µm in diameter This requires extensive folding, which is accomplished by Histones Chromatin Structure is based on Successive Levels of DNA Packing: 1) DNA strand 2) Nucleosomes or “beads on a string” Histones = Small proteins that are rich in basic amino acids and that bind to DNA, forming chromatin. (H1, H2A, H2B, H3, and H4 are used) Partially unfolded chromatin (DNA and its associated proteins) resembles beads spaced along the DNA string. Each beadlike structure is a nucleosome. Nucleosome = The basic unit of DNA packing; it is formed from DNA wound around a protein core that consists of 2 copies each of 4 types of histone. may control gene expression 3) Chromatin Fiber Nucleosomes are zig-zagged into a 3-D structure using H1 proteins 4) Looped Domains Attached to another protein scaffold Euchromatin = loosely attached, allowing access of RNA polymerase and, therefore, transcription 5) Coil and fold further, compacting the chromatin into a mitotic chromosomes characteristic of metaphase Heterochromatin = chromatin that stays in this condensed state even in interphase and is rarely (if ever) transcribed