* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Determination of Amino Acid Sequence
Survey
Document related concepts
Bimolecular fluorescence complementation wikipedia , lookup
Circular dichroism wikipedia , lookup
Degradomics wikipedia , lookup
Protein domain wikipedia , lookup
Protein purification wikipedia , lookup
Protein folding wikipedia , lookup
List of types of proteins wikipedia , lookup
Western blot wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Homology modeling wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Alpha helix wikipedia , lookup
Transcript
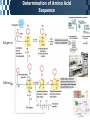
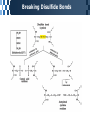
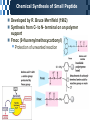
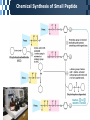
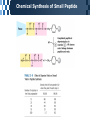
The Covalent Structure of Proteins Determination of Amino Acid Sequence Determination of amino acid sequence from a protein Sanger’s method N-terminal labeling and identification Using FDNB (1-fluoro-2,4-dinitrobenzene) Edman Degradation Sequencing of the entire polypeptide Sequential labeling and removal of the N-terminal amino acid Sequenator Automated sequencing of proteins Accuracy is depending on the efficiency of the individual chemical step > 99% in modern sequenator Translation from DNA sequence DNA sequence protein sequence Protein sequence cloning of the gene Determination of Amino Acid Sequence Sanger Edman Sequencing Large Proteins Breaking disulfide bonds Oxidation by performic acid Reduction and carboxymethylation Cleaving the polypeptide chain Using proteases Cleavage of peptide bond next to particular amino acid residues Trypsin: Lys, Arg Sequencing of peptides Ordering peptide fragments Compare sequences generated from different cleavage methods Locating disulfide bonds Comparison of cleavage fragment with or without breaking disulfide bonds Breaking Disulfide Bonds Sequencing Large Proteins Determination of Amino Acid Sequences Using Mass spectroscopy Components of mass spectrometer Ionizer: converting molecules to gas phase ions Soft ionizer for large molecules MALDI MS ESI MS Mass analyzer: separate the ions according to the m/z Time of flight (TOF) Measuring the time take by ions to travel to the detector Ion detector Mass spectrometer for protein analysis Determination of molecular weight Determination of short polypeptide sequence Tandem MS, MS/MS MALDI-TOF Matrix-assisted laser desorption/ionization mass spectrormetry Protein placed in a light-absorbing matrix Ionization and desorption of proteins by a short pulse of laser ESI-TOF Electrospary ionization mass spectrometry Passing of analyte solution through a charged needle with a high electrical potential Dispersion of charged microdroplets John Fenn(2002) Virginia Commonwealth University MS/MS Chemical Synthesis of Small Peptide Developed by R. Bruce Merrifield (1962) Synthesis from C- to N- terminal on an polymer support Fmoc (9-fluorenylmethoxycarbonyl) Protection of unwanted reaction Chemical Synthesis of Small Peptide Chemical Synthesis of Small Peptide