* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download differential gene expression
Gene desert wikipedia , lookup
Protein moonlighting wikipedia , lookup
Transcription factor wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Epitranscriptome wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Molecular evolution wikipedia , lookup
Community fingerprinting wikipedia , lookup
Non-coding DNA wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Genome evolution wikipedia , lookup
List of types of proteins wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Gene expression profiling wikipedia , lookup
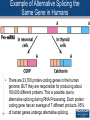
Expression vector wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
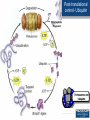
Promoter (genetics) wikipedia , lookup
Gene regulatory network wikipedia , lookup
Gene expression wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
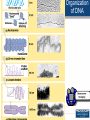
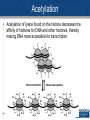
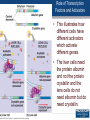
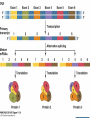
Agenda 3/22 • Stickleback switches video • Stickleback switches model and activity Homework: Gene expression essay (due 3/23) Turn in: Video notes Warm up: How does DNA organization differ in eukaryotes and prokaryotes? 1 • How is eukaryotic gene expression different than prokaryotic gene expression? • How do our cells become different and specialized? 2 Agenda 3/23 • Eukaryotic control lecture • Lactase click and learn • Homework: Development Video and Notes *, Finish Essay • Turn in: Nothing ...unless essay is finished 3 Control of Eukaryotic Gene Expression Eukaryotic Gene Regulation Prokaryotic regulation is different from eukaryotic regulation. 1. Eukaryotic cells have many more genes (23,700 in human cells) in their genomes than prokaryotic cells (average 3000). 2. Physically there are more obstacles as eukaryotic chromatin is wrapped around histone proteins. 3. There are more non-histone proteins that are used in eukaryotic gene expression than in prokaryotic gene expression. 5 Eukaryotic Gene Regulation in Multicellular Organisms • Almost all the cells in an organism are genetically identical or totipotent. • Differences between cell types result from differential gene expression -- the expression of different genes by cells with the same genome. • Errors in gene expression can lead to diseases including cancer. • Gene expression is regulated at many stages. 6 Organization of DNA 7 Three Levels of Control • 1. Pre-transcriptional – Methylation – Acetylation – Transcription factors • 2. Post-transcriptional – Alternative splicing • 3. Post-translational – ubiquitination 8 Epigenetics • Epigenetics refers to processes that influence gene expression or function without changing the underlying DNA sequence. 1. Acetylation 2. Methylation 9 Acetylation • Acetylation of lysine found on the histone decreases the affinity of histones for DNA and other histones, thereby making DNA more accessible for transcription. 10 Methylation • A methyl group can be added to the nitrogenous bases of cytosine that are followed by guanine • Different cells have different methylation patterns, which contributes to the differences in gene expression in different cell types. • Methylation makes DNA less likely to be transcribed 11 12 The role of enhancers and transcription factors. • a. Transcription factors bind to the promoter region (TATA box) for the RNA polymerase to attach. • b. Inducer regions which may help transcription factors to bind. • c. DNA site control elements even further upstream (thousands of nucleotides away) called a enhancers. – DNA enhancers can work by a protein (activator) attaching to the enhancer. – The DNA then loops the DNA back on itself to attach to the promoter region. 13 Role of Transcription Factors 14 Role of Transcription Factors and Activators • This illustrates how different cells have different activators which activate different genes. • The liver cells need the protein albumin and not the protein crystallin and the lens cells do not need albumin but do need crystallin. 15 Post-Transcriptional Control Alternative Splicing Alternative splicing • Once the immature mRNA is made, it could be processed in different ways to give rise to different mature mRNA and thus different proteins. 16 17 Example of Alternative Splicing the Same Gene in Humans • There are 23,700 protein-coding genes in the human genome; BUT they are responsible for producing about 160,000 different proteins. This is possible due to alternative splicing during RNA Processing. Each protein coding gene has an average of 7 different products. 95% of human genes undergo alternative splicing. 18 Post-translational control- Ubiquitin 19