* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Disulfide bridge assignment in complex proteins - HES
Survey
Document related concepts
Structural alignment wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Homology modeling wikipedia , lookup
Protein folding wikipedia , lookup
Circular dichroism wikipedia , lookup
Protein domain wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Protein structure prediction wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein purification wikipedia , lookup
Protein moonlighting wikipedia , lookup
Western blot wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Transcript
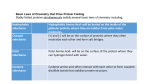
Disulfide bridge assignment in complex proteins Partners HES-SO Valais Collaborator(s) M. Goyder, D. Prim, M. Pfeifer, F. Kálmán Description Assigning disulfide bridges is an important component of the analytical strategy during recombinant protein production, for which mass spectrometry (MS) is an important technique. Venom proteins, such as the threefinger toxins, pose a particular challenge due to their complex arrangement of disulfide linkages. At FTV, we are developing capabilities for straight forward, efficient disulfide bridge assignment in proteins using mass spectrometry, in particular, to enable the study of 'challenging' proteins such as venom proteins, which fail simple disulfide bridge assignment methods. The disulfide assignment strategy is highly dependent on the protein sequence and disulfide bonding pattern. Thus to study a variety of proteins with challenging disulfide connectivities, the toolkit of techniques in this area at FTV are being developed. This includes sample preparation methods, such as chemical/enzymatic digestion and partial reduction and, mass spectrometric techniques (using a QTOF) to fragment along the polypeptide backbone and/or break disulfide bridges, determine with high accuracy the masses and elucidate the disulfide structure. 3D structure of the three-finger toxin, mambin MS/MS based disulfide assignment strategy Highlights of Analytical Sciences in Switzerland – Mass Spectrometric Characterization of Disulfide Bridges in Snake Venom Proteins Miriam S. Goyder, Marc E. Pfeifer and Franka Kálmán (Chimia, 2014, 68(3):184) Strategies in mass spectrometry for the assignment of Cys-Cys disulfide connectivities in proteins Miriam S. Goyder, Fabien Rebeaud, Marc E. Pfeifer and Franka Kálmán (Expert Review in Proteomics, 2013, 10(5):489-501) URL http://itv.hevs.ch/ Contact Franka Kálmán, Ph.D [email protected] T +41 27 606 86 60